Genetic history of Cambridgeshire before and after the Black Death
- PMID: 38232165
- PMCID: PMC10793959
- DOI: 10.1126/sciadv.adi5903
Genetic history of Cambridgeshire before and after the Black Death
Abstract
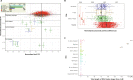
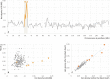
The extent of the devastation of the Black Death pandemic (1346-1353) on European populations is known from documentary sources and its bacterial source illuminated by studies of ancient pathogen DNA. What has remained less understood is the effect of the pandemic on human mobility and genetic diversity at the local scale. Here, we report 275 ancient genomes, including 109 with coverage >0.1×, from later medieval and postmedieval Cambridgeshire of individuals buried before and after the Black Death. Consistent with the function of the institutions, we found a lack of close relatives among the friars and the inmates of the hospital in contrast to their abundance in general urban and rural parish communities. While we detect long-term shifts in local genetic ancestry in Cambridgeshire, we find no evidence of major changes in genetic ancestry nor higher differentiation of immune loci between cohorts living before and after the Black Death.
Figures


References
-
- Skoglund P., Mathieson I., Ancient genomics of modern humans: The first decade. Annu. Rev. Genomics Hum. Genet. 19, 381–404 (2018). - PubMed
-
- Orlando L., Allaby R., Skoglund P., Der Sarkissian C., Stockhammer P. W., Ávila-Arcos M. C., Fu Q., Krause J., Willerslev E., Stone A. C., Warinner C., Ancient DNA analysis. Nat. Rev. Methods Primer 1, 14 (2021).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

