Type 1 immunity enables neonatal thymic ILC1 production
- PMID: 38232171
- PMCID: PMC10793954
- DOI: 10.1126/sciadv.adh5520
Type 1 immunity enables neonatal thymic ILC1 production
Abstract
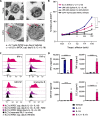
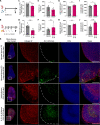
Acute thymic atrophy occurs following type 1 inflammatory conditions such as viral infection and sepsis, resulting in cell death and disruption of T cell development. However, the impact type 1 immunity has on thymic-resident innate lymphoid cells (ILCs) remains unclear. Single-cell RNA sequencing revealed neonatal thymic-resident type 1 ILCs (ILC1s) as a unique and immature subset compared to ILC1s in other primary lymphoid organs. Culturing murine neonatal thymic lobes with the type 1 cytokines interleukin-12 (IL-12) and IL-18 resulted in a rapid expansion and thymic egress of KLRG1+CXCR6+ cytotoxic ILC1s. Live imaging showed the subcapsular thymic localization and exit of ILC1s following IL-12 + IL-18 stimulation. Similarly, murine cytomegalovirus infection in neonates resulted in thymic atrophy and subcapsular localization of thymic-resident ILC1s. Neonatal thymic grafting revealed that type 1 inflammation enhances the homing of cytokine-producing thymus-derived ILC1s to the liver and peritoneal cavity. Together, we show that type 1 immunity promotes the expansion and peripheral homing of thymic-derived ILC1s.
Figures





Similar articles
-
Delta-like 4-Derived Notch Signals Differentially Regulate Thymic Generation of Skin-Homing CCR10+NK1.1+ Innate Lymphoid Cells at Neonatal and Adult Stages.J Immunol. 2022 Sep 1;209(5):950-959. doi: 10.4049/jimmunol.2100870. Epub 2022 Aug 3. J Immunol. 2022. PMID: 35922065 Free PMC article.
-
Liver type 1 innate lymphoid cells lacking IL-7 receptor are a native killer cell subset fostered by parenchymal niches.Elife. 2023 Jun 22;12:e84209. doi: 10.7554/eLife.84209. Elife. 2023. PMID: 37352115 Free PMC article.
-
TL1A and IL-18 synergy promotes GM-CSF-dependent thymic granulopoiesis in mice.Cell Mol Immunol. 2024 Aug;21(8):807-825. doi: 10.1038/s41423-024-01180-8. Epub 2024 Jun 5. Cell Mol Immunol. 2024. PMID: 38839915 Free PMC article.
-
Type 1 innate lymphoid cells: Soldiers at the front line of immunity.Biomed J. 2021 Apr;44(2):115-122. doi: 10.1016/j.bj.2020.10.001. Epub 2020 Nov 19. Biomed J. 2021. PMID: 33839081 Free PMC article. Review.
-
Emerging roles of type 1 innate lymphoid cells in tumour pathogenesis and cancer immunotherapy.Explor Target Antitumor Ther. 2024;5(2):296-315. doi: 10.37349/etat.2024.00219. Epub 2024 Apr 23. Explor Target Antitumor Ther. 2024. PMID: 38745765 Free PMC article. Review.
Cited by
-
The thymus road to a T cell: migration, selection, and atrophy.Front Immunol. 2024 Aug 27;15:1443910. doi: 10.3389/fimmu.2024.1443910. eCollection 2024. Front Immunol. 2024. PMID: 39257583 Free PMC article. Review.
-
Baicalin restores innate lymphoid immune imbalance during exacerbation of COPD.Immunol Res. 2025 Apr 16;73(1):71. doi: 10.1007/s12026-025-09629-2. Immunol Res. 2025. PMID: 40234295 Free PMC article.
-
Reading of human acute immune dynamics in omicron SARS-CoV-2 breakthrough infection.Emerg Microbes Infect. 2025 Dec;14(1):2494705. doi: 10.1080/22221751.2025.2494705. Epub 2025 May 6. Emerg Microbes Infect. 2025. PMID: 40231451 Free PMC article.
-
Spatial mapping of innate lymphoid cells in human lymphoid tissues and lymphoma at single-cell resolution.Nat Commun. 2025 May 15;16(1):4545. doi: 10.1038/s41467-025-59811-1. Nat Commun. 2025. PMID: 40374674 Free PMC article.
References
-
- Gherardi M. M., Ramírez J. C., Esteban M., IL-12 and IL-18 act in synergy to clear vaccinia virus infection: Involvement of innate and adaptive components of the immune system. J. Gen. Virol. 84 (Pt. 8), 1961–1972 (2003). - PubMed
-
- Pien G. C., Satoskar A. R., Takeda K., Akira S., Biron C. A., Cutting edge: Selective IL-18 requirements for induction of compartmental IFN-γ responses during viral infection. J. Immunol. 165, 4787–4791 (2000). - PubMed
-
- Vanden Berghe T., Demon D., Bogaert P., Vandendriessche B., Goethals A., Depuydt B., Vuylsteke M., Roelandt R., Van Wonterghem E., Vandenbroecke J., Choi S. M., Meyer E., Krautwald S., Declercq W., Takahashi N., Cauwels A., Vandenabeele P., Simultaneous targeting of IL-1 and IL-18 is required for protection against inflammatory and septic shock. Am. J. Respir. Crit. Care Med. 189, 282–291 (2014). - PubMed
-
- Mera S., Tatulescu D., Cismaru C., Bondor C., Slavcovici A., Zanc V., Carstina D., Oltean M., Multiplex cytokine profiling in patients with sepsis. APMIS. 119, 155–163 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

