Haplotype-specific assembly of shattered chromosomes in esophageal adenocarcinomas
- PMID: 38232733
- PMCID: PMC10879010
- DOI: 10.1016/j.xgen.2023.100484
Haplotype-specific assembly of shattered chromosomes in esophageal adenocarcinomas
Abstract
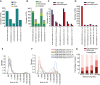
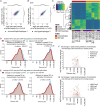
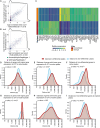
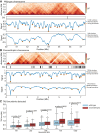
The epigenetic landscape of cancer is regulated by many factors, but primarily it derives from the underlying genome sequence. Chromothripsis is a catastrophic localized genome shattering event that drives, and often initiates, cancer evolution. We characterized five esophageal adenocarcinoma organoids with chromothripsis using long-read sequencing and transcriptome and epigenome profiling. Complex structural variation and subclonal variants meant that haplotype-aware de novo methods were required to generate contiguous cancer genome assemblies. Chromosomes were assembled separately and scaffolded using haplotype-resolved Hi-C reads, producing accurate assemblies even with up to 900 structural rearrangements. There were widespread differences between the chromothriptic and wild-type copies of chromosomes in topologically associated domains, chromatin accessibility, histone modifications, and gene expression. Differential epigenome peaks were most enriched within 10 kb of chromothriptic structural variants. Alterations in transcriptome and higher-order chromosome organization frequently occurred near differential epigenetic marks. Overall, chromothripsis reshapes gene regulation, causing coordinated changes in epigenetic landscape, transcription, and chromosome conformation.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests P.J.C. is an academic co-founder, stockholder, and consultant for Quotient Therapeutics.
Figures




References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

