NMDARs activation regulates endothelial ferroptosis via the PP2A-AMPK-HMGB1 axis
- PMID: 38233385
- PMCID: PMC10794209
- DOI: 10.1038/s41420-023-01794-3
NMDARs activation regulates endothelial ferroptosis via the PP2A-AMPK-HMGB1 axis
Abstract
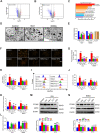
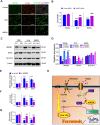
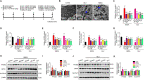
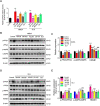
N-methyl-D-aspartate receptors (NMDARs) are ligand-gated, voltage-dependent channels of the ionotropic glutamate receptor family. The present study explored whether NMDAR activation induced ferroptosis in vascular endothelial cells and its complicated mechanisms in vivo and in vitro. Various detection approaches were used to determine the ferroptosis-related cellular iron content, lipid reactive oxygen species (LOS), siRNA molecules, RNA-sequence, MDA, GSH, and western blotting. The AMPK activator Acadesine (AICAR), HMGB1 inhibitor glycyrrhizin (GLY), PP2A inhibitor LB-100, and NMDAR inhibitor MK801 were used to investigate the involved in vivo and in vitro pathways. The activation of NMDAR with L-glutamic acid (GLU) or NMDA significantly promoted cellular ferroptosis, iron content, MDA, and the PTGS2 expression, while decreasing GPX4 expression and GSH concentration in human umbilical vein endothelial cells (HUVECs), which was reversed by ferroptosis inhibitors Ferrostatin-1(Fer-1), Liproxstatin-1 (Lip-1), or Deferoxamine (DFO). RNA-seq revealed that ferroptosis and SLC7A11 participate in NMDA or GLU-mediated NMDAR activation. The PP2A-AMPK-HMGB1 pathway was majorly associated with NMDAR activation-induced ferroptosis, validated using the PP2A inhibitor LB-100, AMPK activator AICAR, or HMGB1 siRNA. The role of NMDAR in ferroptosis was validated in HUVECs induced with the ferroptosis activator errasin or RSL3 and counteracted by the NMDAR inhibitor MK-801. The in vivo results showed that NMDA- or GLU-induced ferroptosis and LOS production was reversed by MK-801, LB-100, AICAR, MK-801, and GLY, confirming that the PP2A-AMPK-HMGB1 pathway is involved in NMDAR activation-induced vascular endothelium ferroptosis. In conclusion, the present study demonstrated a novel role of NMDAR in endothelial cell injury by regulating ferroptosis via the PP2A-AMPK-HMGB1 pathway.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials

