Leveraging single-cell ATAC-seq and RNA-seq to identify disease-critical fetal and adult brain cell types
- PMID: 38233398
- PMCID: PMC10794712
- DOI: 10.1038/s41467-024-44742-0
Leveraging single-cell ATAC-seq and RNA-seq to identify disease-critical fetal and adult brain cell types
Abstract
Prioritizing disease-critical cell types by integrating genome-wide association studies (GWAS) with functional data is a fundamental goal. Single-cell chromatin accessibility (scATAC-seq) and gene expression (scRNA-seq) have characterized cell types at high resolution, and studies integrating GWAS with scRNA-seq have shown promise, but studies integrating GWAS with scATAC-seq have been limited. Here, we identify disease-critical fetal and adult brain cell types by integrating GWAS summary statistics from 28 brain-related diseases/traits (average N = 298 K) with 3.2 million scATAC-seq and scRNA-seq profiles from 83 cell types. We identified disease-critical fetal (respectively adult) brain cell types for 22 (respectively 23) of 28 traits using scATAC-seq, and for 8 (respectively 17) of 28 traits using scRNA-seq. Significant scATAC-seq enrichments included fetal photoreceptor cells for major depressive disorder, fetal ganglion cells for BMI, fetal astrocytes for ADHD, and adult VGLUT2 excitatory neurons for schizophrenia. Our findings improve our understanding of brain-related diseases/traits and inform future analyses.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures
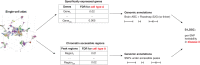
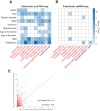
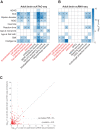
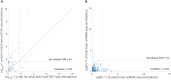
References
MeSH terms
Substances
Grants and funding
- P01 AI148102/AI/NIAID NIH HHS/United States
- R00 HG012203/HG/NHGRI NIH HHS/United States
- R37 MH107649/MH/NIMH NIH HHS/United States
- U01 HG012009/HG/NHGRI NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- F31 HG010818/HG/NHGRI NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 MH115676/MH/NIMH NIH HHS/United States
- U01 HG009379/HG/NHGRI NIH HHS/United States
- R01 MH101244/MH/NIMH NIH HHS/United States
- R01 MH109978/MH/NIMH NIH HHS/United States
- U01 MH119509/MH/NIMH NIH HHS/United States
- R01 HG006399/HG/NHGRI NIH HHS/United States
- R56 HG013083/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources

