METTL3 facilitates renal cell carcinoma progression by PLOD2 m6A-methylation under prolonged hypoxia
- PMID: 38233403
- PMCID: PMC10794171
- DOI: 10.1038/s41419-023-06411-w
METTL3 facilitates renal cell carcinoma progression by PLOD2 m6A-methylation under prolonged hypoxia
Abstract
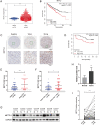
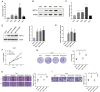
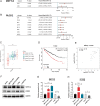
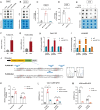
N6-methyladenosine (m6A) is the most prevalent reversible modification in eukaryotic mRNA, and it plays a critical role in tumor progression. The purpose of this study was to investigate the function and regulatory mechanisms of the methyltransferase METTL3 in renal cell carcinoma (RCC). METTL3 expression was upregulated and predicted a poor prognosis in patients with advanced RCC. METTL3 facilitated the proliferation, migration, and invasion of RCC cells, depending on its methylase activity. METTL3 positively regulated the expression of PLOD2, and both genes were triggered under prolonged hypoxia. Mechanistically, hypoxia-induced the binding of HIF-1α to the METTL3 promoter, which enhanced its transcriptional activity. METTL3-mediated m6A modifications of PLOD2 mRNA at 3'UTR region, promoting the translation of PLOD2 protein. Furthermore, silencing METTL3 impaired RCC progression in vitro. In vivo, administration of highly potent and selective METTL3 inhibitor STM2457 showed anti-tumor effects, whereas AAV9-mediated re-transduction of PLOD2 largely abolished the above phenomenon in a subcutaneous mouse model. These findings reveal that hypoxia and HIF-driven METTL3 transcription promote RCC progression by increasing PLOD2 expression in an m6A-dependent manner, suggesting that METTL3 may serve as a novel pharmaceutical intervention for RCC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests. This study was approved by the Ethics Committee of The Third Affiliated Hospital of Soochow University, and written informed consent was obtained from all participants. All procedures performed in studies involving human participants were compliant with ethical standards.
Figures

Similar articles
-
METTL3 and IGF2BP1-Mediated m6A Modification of ZHX2 Promotes Tumor Property of Renal Cell Carcinoma.Kidney Blood Press Res. 2024;49(1):787-798. doi: 10.1159/000540483. Epub 2024 Aug 19. Kidney Blood Press Res. 2024. PMID: 39159608
-
The methyltransferase METTL3 promotes tumorigenesis via mediating HHLA2 mRNA m6A modification in human renal cell carcinoma.J Transl Med. 2022 Jul 6;20(1):298. doi: 10.1186/s12967-022-03496-3. J Transl Med. 2022. PMID: 35794583 Free PMC article.
-
Procollagen-lysine 2-oxoglutarate 5-dioxygenase 2 promotes hypoxia-induced glioma migration and invasion.Oncotarget. 2017 Apr 4;8(14):23401-23413. doi: 10.18632/oncotarget.15581. Oncotarget. 2017. PMID: 28423580 Free PMC article.
-
ZNF677 suppresses renal cell carcinoma progression through N6-methyladenosine and transcriptional repression of CDKN3.Clin Transl Med. 2022 Jun;12(6):e906. doi: 10.1002/ctm2.906. Clin Transl Med. 2022. PMID: 35678231 Free PMC article.
-
PLOD2 in cancer research.Biomed Pharmacother. 2017 Jun;90:670-676. doi: 10.1016/j.biopha.2017.04.023. Epub 2017 Apr 14. Biomed Pharmacother. 2017. PMID: 28415047 Review.
Cited by
-
Methyltransferase-like 3 is a target for the diagnose and therapy of clear cell renal carcinoma.Front Pharmacol. 2025 Apr 17;16:1534655. doi: 10.3389/fphar.2025.1534655. eCollection 2025. Front Pharmacol. 2025. PMID: 40313614 Free PMC article. Review.
-
STM2457 impairs the proliferation of esophageal squamous cell carcinoma by activating DNA damage response through ATM-Chk2 axis.Med Oncol. 2025 Feb 22;42(3):82. doi: 10.1007/s12032-025-02634-3. Med Oncol. 2025. PMID: 39985567
-
TDERS, an exosome RNA-derived signature predicts prognosis and immunotherapeutic response in clear cell renal cell cancer: a multicohort study.J Natl Cancer Cent. 2024 Aug 19;4(4):382-394. doi: 10.1016/j.jncc.2024.07.002. eCollection 2024 Dec. J Natl Cancer Cent. 2024. PMID: 39735439 Free PMC article.
-
Deciphering the interplay between SETD2 mediated H3K36me3 and RNA N6-methyladenosine in clear cell renal cell carcinoma (ccRCC).Epigenetics. 2025 Dec;20(1):2456418. doi: 10.1080/15592294.2025.2456418. Epub 2025 Jan 28. Epigenetics. 2025. PMID: 39874221 Free PMC article.
-
Decoding m6A RNA methylation in kidney disorders: from molecular insights to therapeutic strategies.J Transl Med. 2025 Jul 10;23(1):771. doi: 10.1186/s12967-025-06817-4. J Transl Med. 2025. PMID: 40640897 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

