LncRNA SOX9-AS1 triggers a transcriptional program involved in lipid metabolic reprogramming, cell migration and invasion in triple-negative breast cancer
- PMID: 38233470
- PMCID: PMC10794186
- DOI: 10.1038/s41598-024-51947-2
LncRNA SOX9-AS1 triggers a transcriptional program involved in lipid metabolic reprogramming, cell migration and invasion in triple-negative breast cancer
Abstract
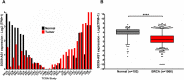
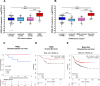
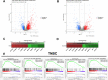
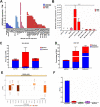
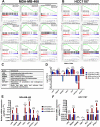
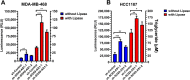
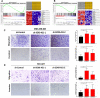
At the molecular level, triple-negative breast cancer (TNBC) is frequently categorized as PAM50 basal-like subtype, but despite the advances in molecular analyses, the clinical outcome for these subtypes is uncertain. Long non-coding RNAs (lncRNAs) are master regulators of genes involved in hallmarks of cancer, which makes them suitable biomarkers for breast cancer (BRCA) diagnosis and prognosis. Here, we evaluated the regulatory role of lncRNA SOX9-AS1 in these subtypes. Using the BRCA-TCGA cohort, we observed that SOX9-AS1 was significantly overexpressed in basal-like and TNBC in comparison with other BRCA subtypes. Survival analyzes showed that SOX9-AS1 overexpression was associated with a favorable prognosis in TNBC and basal-like patients. To study the functions of SOX9-AS1, we determined the expression levels in a panel of nine BRCA cell lines finding increased levels in MDA-MB-468 and HCC1187 TNBC. Using subcellular fractionation in these cell lines, we ascertained that SOX9-AS1 was located in the cytoplasmic compartment. In addition, we performed SOX9-AS1 gene silencing using two short-harping constructs, which were transfected in both cell models and performed a genome-wide RNA-seq analysis. Data showed that 351 lncRNAs and 740 mRNAs were differentially expressed in MDA-MB-468 while 56 lncRNAs and 100 mRNAs were modulated in HCC1187 cells (Log2FC < - 1.5 and > 1.5, p.adj value < 0.05). Pathway analysis revealed that the protein-encoding genes potentially regulate lipid metabolic reprogramming, and epithelial-mesenchymal transition (EMT). Expression of lipid metabolic-related genes LIPE, REEP6, GABRE, FBP1, SCD1, UGT2B11, APOC1 was confirmed by RT-qPCR. Functional analysis demonstrated that the knockdown of SOX9-AS1 increases the triglyceride synthesis, cell migration and invasion in both two TNBC cell lines. In conclusion, high SOX9-AS1 expression predicts an improved clinical course in patients, while the loss of SOX9-AS1 expression enhances the aggressiveness of TNBC cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Garrido-Castro AC, Lin NU, Polyak K. Insights into molecular classifications of triple-negative breast cancer: Improving patient selection for treatment. Cancer Discov. 2019;9:176–198. doi: 10.1158/2159-8290.CD-18-1177. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

