Bacterial community and diversity in the rumen of 11 Mongolian cattle as revealed by 16S rRNA amplicon sequencing
- PMID: 38233488
- PMCID: PMC10794206
- DOI: 10.1038/s41598-024-51828-8
Bacterial community and diversity in the rumen of 11 Mongolian cattle as revealed by 16S rRNA amplicon sequencing
Abstract
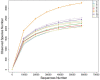
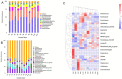
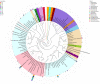
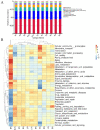
Through microorganism in the rumen of ruminant, plant fiber can be converted to edible food such as meat and milk. Ruminants had a rich and complex microbial community within the rumen, and the bacteria comprised the dominant proportion of the ruminal microbes. High-throughput sequencing offered a viable solution for the study of rumen microbes. In this study, rumen fluid samples were taken from 11 cattle from Inner Mongolian, the DNA of 11 rumen fluid samples were extracted and bacterial amplicons of the V4 regions of 16S rRNA were subjected to Illumina sequencing. More than 90,000 raw reads and 60,000 effect Tags per sample were obtained. 28,122 operational taxonomic units (OTUs) were observed from 11 samples, in average 2557 ± 361 OTUs for each sample. Bacteroidetes (44.41 ± 7.31%), Firmicutes (29.07 ± 3.78%), and Proteobacteria (7.18 ± 5.63%) were the dominant phyla among the bacteria of rumen, accounting for 82%. At the genus level, the highest relative abundance was Prevotella. Their functions were predicted using the Kyoto Encyclopedia of Genes and Genomes (KEGG). The results showed that they included metabolism, genetic information processing, environmental information processing and cellular processes. It explored the bacterial community diversity and composition of the rumen of Mongolian cattle. On the whole, our research showed that there was a high diversity as well as rich bacterial flora function of rumen bacteria in Mongolian cattle. Meanwhile, these findings provided information for further studies on the relationship between the community, diversity, functions of rumen bacteria and the nutritional physiological functions of the host.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Age-dependent variations in rumen bacterial community of Mongolian cattle from weaning to adulthood.BMC Microbiol. 2022 Sep 7;22(1):213. doi: 10.1186/s12866-022-02627-6. BMC Microbiol. 2022. PMID: 36071396 Free PMC article.
-
Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants.J Dairy Sci. 2016 Dec;99(12):9668-9681. doi: 10.3168/jds.2016-11398. Epub 2016 Sep 28. J Dairy Sci. 2016. PMID: 27692708
-
Community structure of the metabolically active rumen bacterial and archaeal communities of dairy cows over the transition period.PLoS One. 2017 Nov 8;12(11):e0187858. doi: 10.1371/journal.pone.0187858. eCollection 2017. PLoS One. 2017. PMID: 29117259 Free PMC article.
-
Composition of bacterial and archaeal communities in the rumen of dromedary camel using cDNA-amplicon sequencing.Int Microbiol. 2020 May;23(2):137-148. doi: 10.1007/s10123-019-00093-1. Epub 2019 Aug 20. Int Microbiol. 2020. PMID: 31432356 Review.
-
Understanding the Diversity and Roles of the Ruminal Microbiome.J Microbiol. 2024 Mar;62(3):217-230. doi: 10.1007/s12275-024-00121-4. Epub 2024 Apr 25. J Microbiol. 2024. PMID: 38662310 Review.
Cited by
-
GnRH Immunocastration in Male Xizang Sheep: Impacts on Rumen Microbiome and Metabolite Profiles for Enhanced Health and Productivity.Animals (Basel). 2024 Oct 11;14(20):2942. doi: 10.3390/ani14202942. Animals (Basel). 2024. PMID: 39457871 Free PMC article.
-
Rumen Bacterial Community Responses to Three DHA Supplements: A Comparative In Vitro Study.Animals (Basel). 2025 Jan 13;15(2):196. doi: 10.3390/ani15020196. Animals (Basel). 2025. PMID: 39858196 Free PMC article.
-
Effects of Licorice Stem and Leaf Forage on Growth and Physiology of Hotan Sheep.Animals (Basel). 2025 May 18;15(10):1459. doi: 10.3390/ani15101459. Animals (Basel). 2025. PMID: 40427336 Free PMC article.
References
-
- Zhou M, Chen Y, Guan LL. Rumen bacteria. In: Puniya A, Singh R, Kamra D, editors. Rumen Microbiology: From Evolution to Revolution. Springer; 2015.
MeSH terms
Substances
Grants and funding
- No. 2020MS03030/Inner Mongolia Autonomous Region Natural Science Foundation Project
- No. 2020MS03030/Inner Mongolia Autonomous Region Natural Science Foundation Project
- No. 2020MS03030/Inner Mongolia Autonomous Region Natural Science Foundation Project
- No. ZTY2023032/Inner Mongolia Autonomous Region Directly Affiliated Universities Basic Research Business Fee Project
- No. ZTY2023032/Inner Mongolia Autonomous Region Directly Affiliated Universities Basic Research Business Fee Project
- No. ZTY2023032/Inner Mongolia Autonomous Region Directly Affiliated Universities Basic Research Business Fee Project
- No. S20231131Z/Inner Mongolia Autonomous Region Graduate Research Innovation Project
- No. S20231131Z/Inner Mongolia Autonomous Region Graduate Research Innovation Project
- No. S20231131Z/Inner Mongolia Autonomous Region Graduate Research Innovation Project
LinkOut - more resources
Full Text Sources

