Targeting protein-protein interaction interfaces with antiviral N protein inhibitor in SARS-CoV-2
- PMID: 38234090
- PMCID: PMC10912909
- DOI: 10.1016/j.bpj.2024.01.013
Targeting protein-protein interaction interfaces with antiviral N protein inhibitor in SARS-CoV-2
Abstract
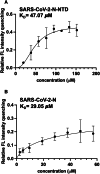
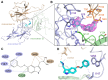
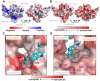
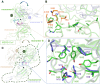
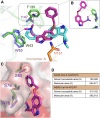
Coronaviruses not only pose significant global public health threats but also cause extensive damage to livestock-based industries. Previous studies have shown that 5-benzyloxygramine (P3) targets the Middle East respiratory syndrome coronavirus (MERS-CoV) nucleocapsid (N) protein N-terminal domain (N-NTD), inducing non-native protein-protein interactions (PPIs) that impair N protein function. Moreover, P3 exhibits broad-spectrum antiviral activity against CoVs. The sequence similarity of N proteins is relatively low among CoVs, further exhibiting notable variations in the hydrophobic residue responsible for non-native PPIs in the N-NTD. Therefore, to ascertain the mechanism by which P3 demonstrates broad-spectrum anti-CoV activity, we determined the crystal structure of the SARS-CoV-2 N-NTD:P3 complex. We found that P3 was positioned in the dimeric N-NTD via hydrophobic contacts. Compared with the interfaces in MERS-CoV N-NTD, P3 had a reversed orientation in SARS-CoV-2 N-NTD. The Phe residue in the MERS-CoV N-NTD:P3 complex stabilized both P3 moieties. However, in the SARS-CoV-2 N-NTD:P3 complex, the Ile residue formed only one interaction with the P3 benzene ring. Moreover, the pocket in the SARS-CoV-2 N-NTD:P3 complex was more hydrophobic, favoring the insertion of the P3 benzene ring into the complex. Nevertheless, hydrophobic interactions remained the primary stabilizing force in both complexes. These findings suggested that despite the differences in the sequence, P3 can accommodate a hydrophobic pocket in N-NTD to mediate a non-native PPI, enabling its effectiveness against various CoVs.
Copyright © 2024 Biophysical Society. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

Similar articles
-
Structure-Based Stabilization of Non-native Protein-Protein Interactions of Coronavirus Nucleocapsid Proteins in Antiviral Drug Design.J Med Chem. 2020 Mar 26;63(6):3131-3141. doi: 10.1021/acs.jmedchem.9b01913. Epub 2020 Mar 11. J Med Chem. 2020. PMID: 32105468
-
Patient-derived monoclonal antibodies to SARS-CoV-2 nucleocapsid protein N-terminal and C-terminal domains cross-react with their counterparts of SARS-CoV, but not other human betacoronaviruses.Front Immunol. 2023 Jan 31;14:1093709. doi: 10.3389/fimmu.2023.1093709. eCollection 2023. Front Immunol. 2023. PMID: 36798118 Free PMC article.
-
Unlocking COVID therapeutic targets: A structure-based rationale against SARS-CoV-2, SARS-CoV and MERS-CoV Spike.Comput Struct Biotechnol J. 2020 Jul 31;18:2117-2131. doi: 10.1016/j.csbj.2020.07.017. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32913581 Free PMC article.
-
Lessons learnt from broad-spectrum coronavirus antiviral drug discovery.Expert Opin Drug Discov. 2024 Sep;19(9):1023-1041. doi: 10.1080/17460441.2024.2385598. Epub 2024 Jul 30. Expert Opin Drug Discov. 2024. PMID: 39078037 Review.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

