Benefits of spatial uncertainty aggregation for segmentation in digital pathology
- PMID: 38234584
- PMCID: PMC10790788
- DOI: 10.1117/1.JMI.11.1.017501
Benefits of spatial uncertainty aggregation for segmentation in digital pathology
Abstract
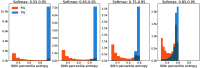
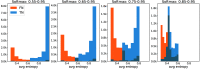
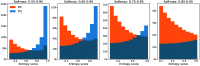
Purpose: Uncertainty estimation has gained significant attention in recent years for its potential to enhance the performance of deep learning (DL) algorithms in medical applications and even potentially address domain shift challenges. However, it is not straightforward to incorporate uncertainty estimation with a DL system to achieve a tangible positive effect. The objective of our work is to evaluate if the proposed spatial uncertainty aggregation (SUA) framework may improve the effectiveness of uncertainty estimation in segmentation tasks. We evaluate if SUA boosts the observed correlation between the uncertainty estimates and false negative (FN) predictions. We also investigate if the observed benefits can translate to tangible improvements in segmentation performance.
Approach: Our SUA framework processes negative prediction regions from a segmentation algorithm and detects FNs based on an aggregated uncertainty score. It can be utilized with many existing uncertainty estimation methods to boost their performance. We compare the SUA framework with a baseline of processing individual pixel's uncertainty independently.
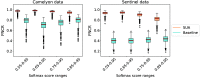
Results: The results demonstrate that SUA is able to detect FN regions. It achieved of 0.92 on the in-domain and 0.85 on the domain-shift test data compared with 0.81 and 0.48 achieved by the baseline uncertainty, respectively. We also demonstrate that SUA yields improved general segmentation performance compared with utilizing the baseline uncertainty.
Conclusions: We propose the SUA framework for incorporating and utilizing uncertainty estimates for FN detection in DL segmentation algorithms for histopathology. The evaluation confirms the benefits of our approach compared with assessing pixel uncertainty independently.
Keywords: computational pathology; deep learning; false negative detection; tumor metastases segmentation; uncertainty estimation.
© 2024 The Authors.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

