This is a preprint.
Genetic variants in DDX53 contribute to Autism Spectrum Disorder associated with the Xp22.11 locus
- PMID: 38234782
- PMCID: PMC10793518
- DOI: 10.1101/2023.12.21.23300383
Genetic variants in DDX53 contribute to Autism Spectrum Disorder associated with the Xp22.11 locus
Update in
-
Genetic variants in DDX53 contribute to autism spectrum disorder associated with the Xp22.11 locus.Am J Hum Genet. 2025 Jan 2;112(1):154-167. doi: 10.1016/j.ajhg.2024.11.003. Epub 2024 Dec 19. Am J Hum Genet. 2025. PMID: 39706195 Free PMC article.
Abstract
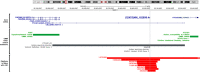
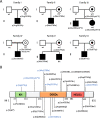
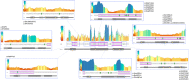
Autism Spectrum Disorder (ASD) exhibits an ~4:1 male-to-female sex bias and is characterized by early-onset impairment of social/communication skills, restricted interests, and stereotyped behaviors. Disruption of the Xp22.11 locus has been associated with ASD in males. This locus includes the three-exon PTCHD1 gene, an adjacent multi-isoform long noncoding RNA (lncRNA) named PTCHD1-AS (spanning ~1Mb), and a poorly characterized single-exon RNA helicase named DDX53 that is intronic to PTCHD1-AS. While the relationship between PTCHD1/PTCHD1-AS and ASD is being studied, the role of DDX53 has not been examined, in part because there is no apparent functional murine orthologue. Through clinical testing, here, we identified 6 males and 1 female with ASD from 6 unrelated families carrying rare, predicted-damaging or loss-of-function variants in DDX53. Then, we examined databases, including the Autism Speaks MSSNG and Simons Foundation Autism Research Initiative, as well as population controls. We identified 24 additional individuals with ASD harboring rare, damaging DDX53 variations, including the same variants detected in two families from the original clinical analysis. In this extended cohort of 31 participants with ASD (28 male, 3 female), we identified 25 mostly maternally-inherited variations in DDX53, including 18 missense changes, 2 truncating variants, 2 in-frame variants, 2 deletions in the 3' UTR and 1 copy number deletion. Our findings in humans support a direct link between DDX53 and ASD, which will be important in clinical genetic testing. These same autism-related findings, coupled with the observation that a functional orthologous gene is not found in mouse, may also influence the design and interpretation of murine-modelling of ASD.
Keywords: Autism; Autism spectrum disorder; DDX53; RNA helicase; Xp22.11 locus.
Conflict of interest statement
Competing interests At the time of this study and its publication, S.W.S. served on the Scientific Advisory Committee of Population Bio. Intellectual property from aspects of his research held at The Hospital for Sick Children are licensed to Athena Diagnostics and Population Bio. These relationships did not influence data interpretation or presentation during this study but are disclosed for potential future considerations. SVM is an employee of GeneDx, LLC. HFP is on the research advisory boards and speaker bureau for Takeda Pharmaceutical, AvroBio, Amicus Therapeutics, Sanofi, Alexion Therapeutics, Denali Therapeutics and Acer Therapeutics. All other authors declare no conflict of interest.
Figures

References
-
- Lai M.C., Lombardo M.V., and Baron-Cohen S. (2014). Autism. Lancet 383, 896–910. - PubMed
-
- Fakhoury M. (2015). Autistic spectrum disorders: A review of clinical features, theories and diagnosis. Int J Dev Neurosci 43, 70–77. - PubMed
-
- Hirota T., and King B.H. (2023). Autism Spectrum Disorder: A Review. JAMA 329, 157–168. - PubMed
-
- Chakrabarti S., and Fombonne E. (2005). Pervasive developmental disorders in preschool children: confirmation of high prevalence. Am J Psychiatry 162, 1133–1141. - PubMed
Web resources
-
- ClinVar; https://www.ncbi.nlm.nih.gov/clinvar
-
- Combined Annotation Dependent Depletion (CADD); http://cadd.gs.washington.edu
-
- Database of Genomic Variants (DGV); http://dgv.tcag.ca/dgv/app/home
-
- DECIPHER; https://decipher.sanger.ac.uk
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
