This is a preprint.
Vaginal Lactobacillus fatty acid response mechanisms reveal a novel strategy for bacterial vaginosis treatment
- PMID: 38234804
- PMCID: PMC10793477
- DOI: 10.1101/2023.12.30.573720
Vaginal Lactobacillus fatty acid response mechanisms reveal a novel strategy for bacterial vaginosis treatment
Update in
-
Vaginal Lactobacillus fatty acid response mechanisms reveal a metabolite-targeted strategy for bacterial vaginosis treatment.Cell. 2024 Sep 19;187(19):5413-5430.e29. doi: 10.1016/j.cell.2024.07.029. Epub 2024 Aug 19. Cell. 2024. PMID: 39163861 Free PMC article.
Abstract
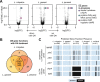
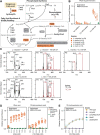
Bacterial vaginosis (BV), a common syndrome characterized by Lactobacillus-deficient vaginal microbiota, is associated with adverse health outcomes. BV often recurs after standard antibiotic therapy in part because antibiotics promote microbiota dominance by Lactobacillus iners instead of Lactobacillus crispatus, which has more beneficial health associations. Strategies to promote L. crispatus and inhibit L. iners are thus needed. We show that oleic acid (OA) and similar long-chain fatty acids simultaneously inhibit L. iners and enhance L. crispatus growth. These phenotypes require OA-inducible genes conserved in L. crispatus and related species, including an oleate hydratase (ohyA) and putative fatty acid efflux pump (farE). FarE mediates OA resistance, while OhyA is robustly active in the human vaginal microbiota and sequesters OA in a derivative form that only ohyA-harboring organisms can exploit. Finally, OA promotes L. crispatus dominance more effectively than antibiotics in an in vitro model of BV, suggesting a novel approach for treatment.
Conflict of interest statement
Conflicts of Interests M.Z., S.M.B., P.C.B., and D.S.K. are co-inventors on a patent related to this work. P.C.B is a co-inventor on patent applications concerning droplet array technologies and serves as a consultant and equity holder of companies in the microfluidics and life sciences industries, including 10x Genomics, GALT/Isolation Bio, Celsius Therapeutics, Next Gen Diagnostics, Cache DNA, Concerto Biosciences, Amber Bio, Stately, Ramona Optics, and Bifrost Biosystems. D.S.K. serves as equity holder of Day Zero Diagnostics.
Figures





Similar articles
-
Vaginal Lactobacillus fatty acid response mechanisms reveal a metabolite-targeted strategy for bacterial vaginosis treatment.Cell. 2024 Sep 19;187(19):5413-5430.e29. doi: 10.1016/j.cell.2024.07.029. Epub 2024 Aug 19. Cell. 2024. PMID: 39163861 Free PMC article.
-
Associations between the vaginal microbiome and Candida colonization in women of reproductive age.Am J Obstet Gynecol. 2020 May;222(5):471.e1-471.e9. doi: 10.1016/j.ajog.2019.10.008. Epub 2019 Oct 22. Am J Obstet Gynecol. 2020. PMID: 31654610 Free PMC article.
-
Cysteine dependence of Lactobacillus iners is a potential therapeutic target for vaginal microbiota modulation.Nat Microbiol. 2022 Mar;7(3):434-450. doi: 10.1038/s41564-022-01070-7. Epub 2022 Mar 3. Nat Microbiol. 2022. PMID: 35241796 Free PMC article.
-
Beyond bacterial vaginosis: vaginal lactobacilli and HIV risk.Microbiome. 2021 Dec 10;9(1):239. doi: 10.1186/s40168-021-01183-x. Microbiome. 2021. PMID: 34893070 Free PMC article. Review.
-
[Characteristics and physiologic role of female lower genital microbiome].Orv Hetil. 2023 Jun 18;164(24):923-930. doi: 10.1556/650.2023.32791. Print 2023 Jun 18. Orv Hetil. 2023. PMID: 37330978 Review. Hungarian.
References
-
- Moreno I., Codoñer F.M., Vilella F., Valbuena D., Martinez-Blanch J.F., Jimenez-Almazán J., Alonso R., Alamá P., Remohí J., Pellicer A., et al. (2016). Evidence that the endometrial microbiota has an effect on implantation success or failure. Am. J. Obstet. Gynecol. 215, 684–703. - PubMed
-
- Ravel J., Moreno I., and Simón C. (2021). Bacterial vaginosis and its association with infertility, endometritis, and pelvic inflammatory disease. Am. J. Obstet. Gynecol. 224, 251–257. - PubMed
-
- Moore D.E., Soules M.R., Klein N.A., Fujimoto V.Y., Agnew K.J., and Eschenbach D.A. (2000). Bacteria in the transfer catheter tip influence the live-birth rate after in vitro fertilization. Fertil. Steril. 74, 1118–1124. - PubMed
-
- Gaudoin M., Rekha P., Morris A., Lynch J., and Acharya U. (1999). Bacterial vaginosis and past chlamydial infection are strongly and independently associated with tubal infertility but do not affect in vitro fertilization success rates. Fertil. Steril. 72, 730–732. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
