This is a preprint.
Long non-coding RNA Malat1 fine-tunes bone homeostasis and repair by orchestrating cellular crosstalk and the β-catenin-OPG/Jagged1 pathway
- PMID: 38234849
- PMCID: PMC10793491
- DOI: 10.21203/rs.3.rs-3793919/v3
Long non-coding RNA Malat1 fine-tunes bone homeostasis and repair by orchestrating cellular crosstalk and the β-catenin-OPG/Jagged1 pathway
Update in
-
Long non-coding RNA Malat1 fine-tunes bone homeostasis and repair by orchestrating cellular crosstalk and β-catenin-OPG/Jagged1 pathway.Elife. 2024 Dec 23;13:RP98900. doi: 10.7554/eLife.98900. Elife. 2024. PMID: 39714456 Free PMC article.
Abstract
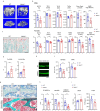
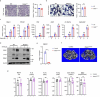
The IncRNA Malat1 was initially believed to be dispensable for physiology due to the lack of observable phenotypes in Malat1 knockout (KO) mice. However, our study challenges this conclusion. We found that both Malat1 KO and conditional KO mice in the osteoblast lineage exhibit significant osteoporosis. Mechanistically, Malat1 acts as an intrinsic regulator in osteoblasts to promote osteogenesis. Interestingly, Malat1 does not directly affect osteoclastogenesis but inhibits osteoclastogenesis in a non-autonomous manner in vivo via integrating crosstalk between multiple cell types, including osteoblasts, osteoclasts and chondrocytes. Our findings substantiate the existence of a novel remodeling network in which Malat1 serves as a central regulator by binding to β-catenin and functioning through the β-catenin-OPG/Jagged1 pathway in osteoblasts and chondrocytes. In pathological conditions, Malat1 significantly promotes bone regeneration in fracture healing. Bone homeostasis and regeneration are crucial to well-being. Our discoveries establish a previous unrecognized paradigm model of Malat1 function in the skeletal system, providing novel mechanistic insights into how a lncRNA integrates cellular crosstalk and molecular networks to fine tune tissue homeostasis, remodeling and repair.
Keywords: Malat1; bone; fracture; homeostasis; lncRNA; long non-coding RNA; remodeling.
Conflict of interest statement
Competing Interests statement The authors have no conflict of interests.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

