Temporal σB stress-response profiles impact Bacillus subtilis fitness
- PMID: 38236030
- PMCID: PMC10900884
- DOI: 10.1128/msphere.00719-23
Temporal σB stress-response profiles impact Bacillus subtilis fitness
Abstract
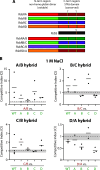
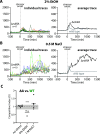
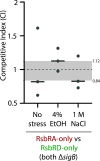
The Gram-positive model organism Bacillus subtilis responds to environmental stressors by activating the alternative sigma factor σB. The sensing apparatus upstream of σB activation is thought to consist of cytoplasmic stressosomes-megadalton-sized protein complexes that include five paralogous proteins known as RsbRs. The RsbRs are presumed to be involved in stress sensing and the subsequent response. Perturbations to the RsbR complement in stressosomes by engineering cells that produce only one of the RsbR paralogs ("single-RsbR strains") lead to altered σB response dynamics with respect to timing and magnitude. Here, we asked whether such changes to σB response dynamics impact the relative fitness of a strain. We competed strain pairs with different RsbR complements under ethanol and sodium chloride stress and found not only differences in relative fitness among wild-type and single-RsbR strains but also different relative fitness values in the two different stressors. We found that the presence of RsbRA, which dominates the wild-type σB response, enhances fitness in ethanol but is detrimental to fitness in NaCl. Meanwhile, RsbRD-only cells were among the most fit in NaCl. Strains producing hybrid RsbR fusion proteins displayed different fitness values that depended on the RsbR proteins from which they were derived. Our results here suggest that σB response dynamics can impact fitness, highlighting the physiological importance of the unusual stressosome-based general stress response system of B. subtilis.
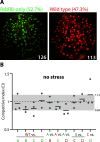
Importance: The model bacterium Bacillus subtilis uses cytoplasmic multiprotein complexes, termed stressosomes, to activate the alternative sigma factor σB when facing environmental stresses. We have previously shown that genetically manipulating the complement of putative sensor proteins in stressosomes can alter the dynamics of the σB response in terms of its magnitude and timing. However, it is unknown whether these response dynamics impact the fitness of cells challenged by environmental stressors. Here, we examine the fitness of strains with different σB responses by competing strain pairs in exponential-phase co-cultures under environmental stress. We find that strains with different response dynamics show different competitive indices that differ by stressor. These results suggest that the dynamics of the σB response can affect the fitness of cells facing environmental stress, highlighting the relevance of different σB dynamics.
Keywords: Bacillus subtilis; competition; fitness; sigma factors; stress response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

