Mind the gap: Epigenetic regulation of chromatin accessibility in plants
- PMID: 38236303
- PMCID: PMC10980423
- DOI: 10.1093/plphys/kiae024
Mind the gap: Epigenetic regulation of chromatin accessibility in plants
Abstract
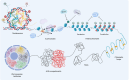
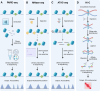
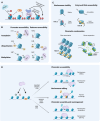
Chromatin plays a crucial role in genome compaction and is fundamental for regulating multiple nuclear processes. Nucleosomes, the basic building blocks of chromatin, are central in regulating these processes, determining chromatin accessibility by limiting access to DNA for various proteins and acting as important signaling hubs. The association of histones with DNA in nucleosomes and the folding of chromatin into higher-order structures are strongly influenced by a variety of epigenetic marks, including DNA methylation, histone variants, and histone post-translational modifications. Additionally, a wide array of chaperones and ATP-dependent remodelers regulate various aspects of nucleosome biology, including assembly, deposition, and positioning. This review provides an overview of recent advances in our mechanistic understanding of how nucleosomes and chromatin organization are regulated by epigenetic marks and remodelers in plants. Furthermore, we present current technologies for profiling chromatin accessibility and organization.
© The Author(s) 2024. Published by Oxford University Press on behalf of American Society of Plant Biologists.
Conflict of interest statement
Conflict of interest statement. None declared.
Figures
Similar articles
-
Deposition and eviction of histone variants define functional chromatin states in plants.Curr Opin Plant Biol. 2022 Oct;69:102266. doi: 10.1016/j.pbi.2022.102266. Epub 2022 Aug 15. Curr Opin Plant Biol. 2022. PMID: 35981458 Review.
-
A complex interplay between histone variants and DNA methylation.J Exp Bot. 2025 Jun 17;76(9):2468-2477. doi: 10.1093/jxb/eraf030. J Exp Bot. 2025. PMID: 39866056 Review.
-
Histone modification: cause or cog?Trends Genet. 2011 Oct;27(10):389-96. doi: 10.1016/j.tig.2011.06.006. Epub 2011 Jul 20. Trends Genet. 2011. PMID: 21764166
-
A comprehensive view of the epigenetic landscape. Part II: Histone post-translational modification, nucleosome level, and chromatin regulation by ncRNAs.Neurotox Res. 2015 Feb;27(2):172-97. doi: 10.1007/s12640-014-9508-6. Epub 2014 Dec 17. Neurotox Res. 2015. PMID: 25516120 Free PMC article. Review.
-
Interplay between histone variants and chaperones in plants.Curr Opin Plant Biol. 2024 Aug;80:102551. doi: 10.1016/j.pbi.2024.102551. Epub 2024 May 21. Curr Opin Plant Biol. 2024. PMID: 38776573 Review.
Cited by
-
Almond Grafting for Plum Pox Virus Resistance Triggers Significant Transcriptomic and Epigenetic Shifts in Peaches.Int J Mol Sci. 2024 Dec 30;26(1):248. doi: 10.3390/ijms26010248. Int J Mol Sci. 2024. PMID: 39796109 Free PMC article.
-
H3K4me3 binding ALFIN-LIKE proteins recruit SWR1 for gene-body deposition of H2A.Z.Genome Biol. 2025 May 21;26(1):137. doi: 10.1186/s13059-025-03605-7. Genome Biol. 2025. PMID: 40399998 Free PMC article.
-
Focus on epigenetics.Plant Physiol. 2024 Mar 29;194(4):1925-1928. doi: 10.1093/plphys/kiae104. Plant Physiol. 2024. PMID: 38401162 Free PMC article. No abstract available.
-
The generation of novel epialleles in plants: the prospective behind re-shaping the epigenome.Front Plant Sci. 2025 Mar 21;16:1544744. doi: 10.3389/fpls.2025.1544744. eCollection 2025. Front Plant Sci. 2025. PMID: 40190658 Free PMC article. Review.
-
Paulownia Witches' Broom Disease: A Comprehensive Review.Microorganisms. 2024 Apr 28;12(5):885. doi: 10.3390/microorganisms12050885. Microorganisms. 2024. PMID: 38792713 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

