Tumor-educated Gr1+CD11b+ cells drive breast cancer metastasis via OSM/IL-6/JAK-induced cancer cell plasticity
- PMID: 38236642
- PMCID: PMC10940099
- DOI: 10.1172/JCI166847
Tumor-educated Gr1+CD11b+ cells drive breast cancer metastasis via OSM/IL-6/JAK-induced cancer cell plasticity
Abstract
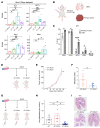
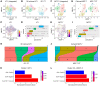
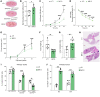
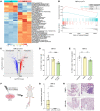
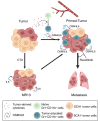
Cancer cell plasticity contributes to therapy resistance and metastasis, which represent the main causes of cancer-related death, including in breast cancer. The tumor microenvironment drives cancer cell plasticity and metastasis, and unraveling the underlying cues may provide novel strategies for managing metastatic disease. Using breast cancer experimental models and transcriptomic analyses, we show that stem cell antigen-1 positive (SCA1+) murine breast cancer cells enriched during tumor progression and metastasis had higher in vitro cancer stem cell-like properties, enhanced in vivo metastatic ability, and generated tumors rich in Gr1hiLy6G+CD11b+ cells. In turn, tumor-educated Gr1+CD11b+ (Tu-Gr1+CD11b+) cells rapidly and transiently converted low metastatic SCA1- cells into highly metastatic SCA1+ cells via secreted oncostatin M (OSM) and IL-6. JAK inhibition prevented OSM/IL-6-induced SCA1+ population enrichment, while OSM/IL-6 depletion suppressed Tu-Gr1+CD11b+-induced SCA1+ population enrichment in vitro and metastasis in vivo. Moreover, chemotherapy-selected highly metastatic 4T1 cells maintained high SCA1+ positivity through autocrine IL-6 production, and in vitro JAK inhibition blunted SCA1 positivity and metastatic capacity. Importantly, Tu-Gr1+CD11b+ cells invoked a gene signature in tumor cells predicting shorter overall survival (OS), relapse-free survival (RFS), and lung metastasis in breast cancer patients. Collectively, our data identified OSM/IL-6/JAK as a clinically relevant paracrine/autocrine axis instigating breast cancer cell plasticity and triggering metastasis.
Keywords: Breast cancer; Oncology.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

