The use of single-cell RNA-seq to study heterogeneity at varying levels of virus-host interactions
- PMID: 38236826
- PMCID: PMC10796064
- DOI: 10.1371/journal.ppat.1011898
The use of single-cell RNA-seq to study heterogeneity at varying levels of virus-host interactions
Abstract
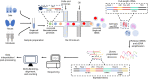
The outcome of viral infection depends on the diversity of the infecting viral population and the heterogeneity of the cell population that is infected. Until almost a decade ago, the study of these dynamic processes during viral infection was challenging and limited to certain targeted measurements. Presently, with the use of single-cell sequencing technology, the complex interface defined by the interactions of cells with infecting virus can now be studied across the breadth of the transcriptome in thousands of individual cells simultaneously. In this review, we will describe the use of single-cell RNA sequencing (scRNA-seq) to study the heterogeneity of viral infections, ranging from individual virions to the immune response between infected individuals. In addition, we highlight certain key experimental limitations and methodological decisions that are critical to analyzing scRNA-seq data at each scale.
Copyright: © 2024 Swaminath, Russell. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

