Priming agents transiently reduce the clearance of cell-free DNA to improve liquid biopsies
- PMID: 38236959
- PMCID: PMC11529396
- DOI: 10.1126/science.adf2341
Priming agents transiently reduce the clearance of cell-free DNA to improve liquid biopsies
Abstract
Liquid biopsies enable early detection and monitoring of diseases such as cancer, but their sensitivity remains limited by the scarcity of analytes such as cell-free DNA (cfDNA) in blood. Improvements to sensitivity have primarily relied on enhancing sequencing technology ex vivo. We sought to transiently augment the level of circulating tumor DNA (ctDNA) in a blood draw by attenuating its clearance in vivo. We report two intravenous priming agents given 1 to 2 hours before a blood draw to recover more ctDNA. Our priming agents consist of nanoparticles that act on the cells responsible for cfDNA clearance and DNA-binding antibodies that protect cfDNA. In tumor-bearing mice, they greatly increase the recovery of ctDNA and improve the sensitivity for detecting small tumors.
Conflict of interest statement
Figures


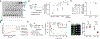
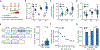
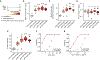
Comment in
-
Improving liquid biopsy for cancer detection.Nat Rev Drug Discov. 2024 Mar;23(3):174. doi: 10.1038/d41573-024-00025-3. Nat Rev Drug Discov. 2024. PMID: 38326470 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

