Gatad2b, associated with the neurodevelopmental syndrome GAND, plays a critical role in neurodevelopment and cortical patterning
- PMID: 38238293
- PMCID: PMC10796954
- DOI: 10.1038/s41398-023-02678-x
Gatad2b, associated with the neurodevelopmental syndrome GAND, plays a critical role in neurodevelopment and cortical patterning
Abstract
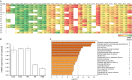
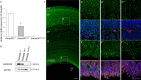
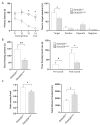
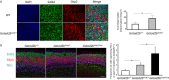
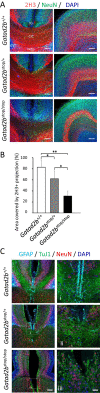
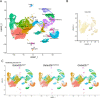
GATAD2B (GATA zinc finger domain containing 2B) variants are associated with the neurodevelopmental syndrome GAND, characterized by intellectual disability (ID), infantile hypotonia, apraxia of speech, epilepsy, macrocephaly and distinct facial features. GATAD2B encodes for a subunit of the Nucleosome Remodeling and Histone Deacetylase (NuRD) complex. NuRD controls transcriptional programs critical for proper neurodevelopment by coupling histone deacetylase with ATP-dependent chromatin remodeling activity. To study mechanisms of pathogenesis for GAND, we characterized a mouse model harboring an inactivating mutation in Gatad2b. Homozygous Gatad2b mutants die perinatally, while haploinsufficient Gatad2b mice exhibit behavioral abnormalities resembling the clinical features of GAND patients. We also observed abnormal cortical patterning, and cellular proportions and cell-specific alterations in the developmental transcriptome in these mice. scRNAseq of embryonic cortex indicated misexpression of genes key for corticogenesis and associated with neurodevelopmental syndromes such as Bcl11b, Nfia and H3f3b and Sox5. These data suggest a crucial role for Gatad2b in brain development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests
Figures
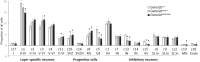
References
-
- Allen Institute for Brain Science, Sestan N, Gerstein MB, Knowles JA, Levitt P, Fischl B, et al. BrainSpan Atlas of the Developing Human Brain. 2023; Available from: http://brainspan.org.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

