Clinical application of tumour-in-normal contamination assessment from whole genome sequencing
- PMID: 38238294
- PMCID: PMC10796348
- DOI: 10.1038/s41467-023-44158-2
Clinical application of tumour-in-normal contamination assessment from whole genome sequencing
Abstract
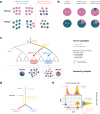
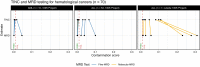
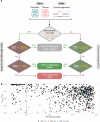
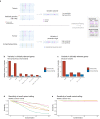
The unexpected contamination of normal samples with tumour cells reduces variant detection sensitivity, compromising downstream analyses in canonical tumour-normal analyses. Leveraging whole-genome sequencing data available at Genomics England, we develop a tool for normal sample contamination assessment, which we validate in silico and against minimal residual disease testing. From a systematic review of [Formula: see text] patients with haematological malignancies and sarcomas, we find contamination across a range of cancer clinical indications and DNA sources, with highest prevalence in saliva samples from acute myeloid leukaemia patients, and sorted CD3+ T-cells from myeloproliferative neoplasms. Further exploration reveals 108 hotspot mutations in genes associated with haematological cancers at risk of being subtracted by standard variant calling pipelines. Our work highlights the importance of contamination assessment for accurate somatic variants detection in research and clinical settings, especially with large-scale sequencing projects being utilised to deliver accurate data from which to make clinical decisions for patient care.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

