Altered DNA methylation within DNMT3A, AHRR, LTA/TNF loci mediates the effect of smoking on inflammatory bowel disease
- PMID: 38238335
- PMCID: PMC10796384
- DOI: 10.1038/s41467-024-44841-y
Altered DNA methylation within DNMT3A, AHRR, LTA/TNF loci mediates the effect of smoking on inflammatory bowel disease
Abstract
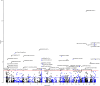
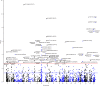
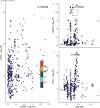
This work aims to investigate how smoking exerts effect on the development of inflammatory bowel disease (IBD). A prospective cohort study and a Mendelian randomization study are first conducted to evaluate the association between smoking behaviors, smoking-related DNA methylation and the risks of Crohn's disease (CD) and ulcerative colitis (UC). We then perform both genome-wide methylation analysis and co-localization analysis to validate the observed associations. Compared to never smoking, current and previous smoking habits are associated with increased CD (P = 7.09 × 10-10) and UC (P < 2 × 10-16) risk, respectively. DNA methylation alteration at cg17742416 [DNMT3A] is linked to both CD (P = 7.30 × 10-8) and UC (P = 1.04 × 10-4) risk, while cg03599224 [LTA/TNF] is associated with CD risk (P = 1.91 × 10-6), and cg14647125 [AHRR] and cg23916896 [AHRR] are linked to UC risk (P = 0.001 and 0.002, respectively). Our study identifies biological mechanisms and pathways involved in the effects of smoking on the pathogenesis of IBD.
© 2024. The Author(s).
Conflict of interest statement
J.K.N. declares to be funded by a grant from the Biocodex Microbiota Foundation. The remaining authors declare no competing interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

