Simulated digestions of free oligosaccharides and mucin-type O-glycans reveal a potential role for Clostridium perfringens
- PMID: 38238389
- PMCID: PMC10796942
- DOI: 10.1038/s41598-023-51012-4
Simulated digestions of free oligosaccharides and mucin-type O-glycans reveal a potential role for Clostridium perfringens
Abstract
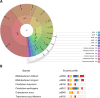
The development of a stable human gut microbiota occurs within the first year of life. Many open questions remain about how microfloral species are influenced by the composition of milk, in particular its content of human milk oligosaccharides (HMOs). The objective is to investigate the effect of the human HMO glycome on bacterial symbiosis and competition, based on the glycoside hydrolase (GH) enzyme activities known to be present in microbial species. We extracted from UniProt a list of all bacterial species catalysing glycoside hydrolase activities (EC 3.2.1.-), cross-referencing with the BRENDA database, and obtained a set of taxonomic lineages and CAZy family data. A set of 13 documented enzyme activities was selected and modelled within an enzyme simulator according to a method described previously in the context of biosynthesis. A diverse population of experimentally observed HMOs was fed to the simulator, and the enzymes matching specific bacterial species were recorded, based on their appearance of individual enzymes in the UniProt dataset. Pairs of bacterial species were identified that possessed complementary enzyme profiles enabling the digestion of the HMO glycome, from which potential symbioses could be inferred. Conversely, bacterial species having similar GH enzyme profiles were considered likely to be in competition for the same set of dietary HMOs within the gut of the newborn. We generated a set of putative biodegradative networks from the simulator output, which provides a visualisation of the ability of organisms to digest HMO and mucin-type O-glycans. B. bifidum, B. longum and C. perfringens species were predicted to have the most diverse GH activity and therefore to excel in their ability to digest these substrates. The expected cooperative role of Bifidobacteriales contrasts with the surprising capacities of the pathogen. These findings indicate that potential pathogens may associate in human gut based on their shared glycoside hydrolase digestive apparatus, and which, in the event of colonisation, might result in dysbiosis. The methods described can readily be adapted to other enzyme categories and species as well as being easily fine-tuneable if new degrading enzymes are identified and require inclusion in the model.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

