Redox signalling regulates breast cancer metastasis via phenotypic and metabolic reprogramming due to p63 activation by HIF1α
- PMID: 38238426
- PMCID: PMC10951347
- DOI: 10.1038/s41416-023-02522-5
Redox signalling regulates breast cancer metastasis via phenotypic and metabolic reprogramming due to p63 activation by HIF1α
Abstract
Background: Redox signaling caused by knockdown (KD) of Glutathione Peroxidase 2 (GPx2) in the PyMT mammary tumour model promotes metastasis via phenotypic and metabolic reprogramming. However, the tumour cell subpopulations and transcriptional regulators governing these processes remained unknown.
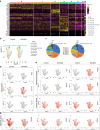
Methods: We used single-cell transcriptomics to decipher the tumour cell subpopulations stimulated by GPx2 KD in the PyMT mammary tumour and paired pulmonary metastases. We analyzed the EMT spectrum across the various tumour cell clusters using pseudotime trajectory analysis and elucidated the transcriptional and metabolic regulation of the hybrid EMT state.
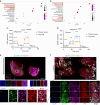
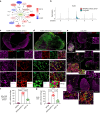
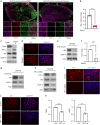
Results: Integration of single-cell transcriptomics between the PyMT/GPx2 KD primary tumour and paired lung metastases unraveled a basal/mesenchymal-like cluster and several luminal-like clusters spanning an EMT spectrum. Interestingly, the luminal clusters at the primary tumour gained mesenchymal gene expression, resulting in epithelial/mesenchymal subpopulations fueled by oxidative phosphorylation (OXPHOS) and glycolysis. By contrast, at distant metastasis, the basal/mesenchymal-like cluster gained luminal and mesenchymal gene expression, resulting in a hybrid subpopulation using OXPHOS, supporting adaptive plasticity. Furthermore, p63 was dramatically upregulated in all hybrid clusters, implying a role in regulating partial EMT and MET at primary and distant sites, respectively. Importantly, these effects were reversed by HIF1α loss or GPx2 gain of function, resulting in metastasis suppression.
Conclusions: Collectively, these results underscored a dramatic effect of redox signaling on p63 activation by HIF1α, underlying phenotypic and metabolic plasticity leading to mammary tumour metastasis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

