Microglia regulate sleep through calcium-dependent modulation of norepinephrine transmission
- PMID: 38238430
- PMCID: PMC10849959
- DOI: 10.1038/s41593-023-01548-5
Microglia regulate sleep through calcium-dependent modulation of norepinephrine transmission
Abstract
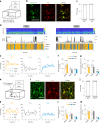
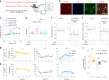
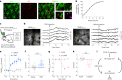
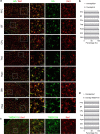
Sleep interacts reciprocally with immune system activity, but its specific relationship with microglia-the resident immune cells in the brain-remains poorly understood. Here, we show in mice that microglia can regulate sleep through a mechanism involving Gi-coupled GPCRs, intracellular Ca2+ signaling and suppression of norepinephrine transmission. Chemogenetic activation of microglia Gi signaling strongly promoted sleep, whereas pharmacological blockade of Gi-coupled P2Y12 receptors decreased sleep. Two-photon imaging in the cortex showed that P2Y12-Gi activation elevated microglia intracellular Ca2+, and blockade of this Ca2+ elevation largely abolished the Gi-induced sleep increase. Microglia Ca2+ level also increased at natural wake-to-sleep transitions, caused partly by reduced norepinephrine levels. Furthermore, imaging of norepinephrine with its biosensor in the cortex showed that microglia P2Y12-Gi activation significantly reduced norepinephrine levels, partly by increasing the adenosine concentration. These findings indicate that microglia can regulate sleep through reciprocal interactions with norepinephrine transmission.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

