KARR-seq reveals cellular higher-order RNA structures and RNA-RNA interactions
- PMID: 38238480
- PMCID: PMC11255127
- DOI: 10.1038/s41587-023-02109-8
KARR-seq reveals cellular higher-order RNA structures and RNA-RNA interactions
Abstract
RNA fate and function are affected by their structures and interactomes. However, how RNA and RNA-binding proteins (RBPs) assemble into higher-order structures and how RNA molecules may interact with each other to facilitate functions remain largely unknown. Here we present KARR-seq, which uses N3-kethoxal labeling and multifunctional chemical crosslinkers to covalently trap and determine RNA-RNA interactions and higher-order RNA structures inside cells, independent of local protein binding to RNA. KARR-seq depicts higher-order RNA structure and detects widespread intermolecular RNA-RNA interactions with high sensitivity and accuracy. Using KARR-seq, we show that translation represses mRNA compaction under native and stress conditions. We determined the higher-order RNA structures of respiratory syncytial virus (RSV) and vesicular stomatitis virus (VSV) and identified RNA-RNA interactions between the viruses and the host RNAs that potentially regulate viral replication.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The University of Chicago has filed a patent application on KARR-seq. C.H. is a scientific founder, a member of the scientific advisory board and an equity holder of Aferna Green, Inc. and AccuaDX, Inc., and is a scientific co-founder and equity holder of Accent Therapeutics, Inc. T.W. is an equity holder of AccuaDX, Inc. The other authors declare no competing interests.
Figures



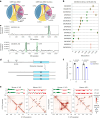
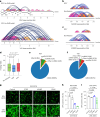
References
-
- Hafner, M. et al. CLIP and complementary methods. Nat. Rev. Methods Primers1, 20 (2021).
-
- Sharma, E., Sterne-Weiler, T., O’Hanlon, D. & Blencowe, B. J. Global mapping of human RNA–RNA interactions. Mol. Cell62, 618–626 (2016). - PubMed
MeSH terms
Substances
Grants and funding
- RM1HG008935/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)
- RM1 HG008935/HG/NHGRI NIH HHS/United States
- R35GM124998/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01HG012780/U.S. Department of Health & Human Services | NIH | National Human Genome Research Institute (NHGRI)
- R01 HG012780/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources

