Common Features of Environmental Mycobacterium chelonae from Colorado Using Partial and Whole Genomic Sequence Analyses
- PMID: 38238596
- PMCID: PMC10796651
- DOI: 10.1007/s00284-023-03589-2
Common Features of Environmental Mycobacterium chelonae from Colorado Using Partial and Whole Genomic Sequence Analyses
Abstract
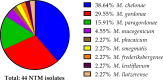
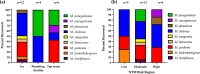
Nontuberculous mycobacteria (NTM) are environmentally acquired opportunistic pathogens that cause chronic lung disease in susceptible individuals. While presumed to be ubiquitous in built and natural environments, NTM environmental studies are limited. While environmental sampling campaigns have been performed in geographic areas of high NTM disease burden, NTM species diversity is less defined among areas of lower disease burden like Colorado. In Colorado, metals such as molybdenum have been correlated with increased risk for NTM infection, yet environmental NTM species diversity has not yet been widely studied. Based on prior regression modeling, three areas of predicted high, moderate, and low NTM risk were identified for environmental sampling in Colorado. Ice, plumbing biofilms, and sink tap water samples were collected from publicly accessible freshwater sources. All samples were microbiologically cultured and NTM were identified using partial rpoB gene sequencing. From these samples, areas of moderate risk were more likely to be NTM positive. NTM recovery from ice was more common than recovery from plumbing biofilms or tap water. Overall, nine different NTM species were identified, including clinically important Mycobacterium chelonae. MinION technology was used to whole genome sequence and compare mutational differences between six M. chelonae genomes, representing three environmental isolates from this study and three other M. chelonae isolates from other sources. Drug resistance genes and prophages were common findings among environmentally derived M. chelonae, promoting the need for expanded environmental sampling campaigns to improve our current understanding of NTM species abundance while opening new avenues for improved targeted drug therapies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest or relevant financial interests to disclose. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
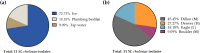


References
-
- Katoch VM. Infections due to non-tuberculous mycobacteria (NTM) Indian J Med Res. 2004;120(4):290–304. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

