Genomic insights into Plasmodium vivax population structure and diversity in central Africa
- PMID: 38238806
- PMCID: PMC10797969
- DOI: 10.1186/s12936-024-04852-y
Genomic insights into Plasmodium vivax population structure and diversity in central Africa
Abstract
Background: Though Plasmodium vivax is the second most common malaria species to infect humans, it has not traditionally been considered a major human health concern in central Africa given the high prevalence of the human Duffy-negative phenotype that is believed to prevent infection. Increasing reports of asymptomatic and symptomatic infections in Duffy-negative individuals throughout Africa raise the possibility that P. vivax is evolving to evade host resistance, but there are few parasite samples with genomic data available from this part of the world.
Methods: Whole genome sequencing of one new P. vivax isolate from the Democratic Republic of the Congo (DRC) was performed and used in population genomics analyses to assess how this central African isolate fits into the global context of this species.
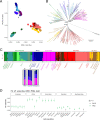
Results: Plasmodium vivax from DRC is similar to other African populations and is not closely related to the non-human primate parasite P. vivax-like. Evidence is found for a duplication of the gene PvDBP and a single copy of PvDBP2.
Conclusion: These results suggest an endemic P. vivax population is present in central Africa. Intentional sampling of P. vivax across Africa would further contextualize this sample within African P. vivax diversity and shed light on the mechanisms of infection in Duffy negative individuals. These results are limited by the uncertainty of how representative this single sample is of the larger population of P. vivax in central Africa.
Keywords: Central Africa; Duffy negative; Genome; Malaria; Plasmodium vivax; Sub-Saharan Africa.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

