Comprehensive full genome analysis of norovirus strains from eastern India, 2017-2021
- PMID: 38238807
- PMCID: PMC10797879
- DOI: 10.1186/s13099-023-00594-5
Comprehensive full genome analysis of norovirus strains from eastern India, 2017-2021
Abstract
Background: Worldwide, noroviruses are the leading cause of acute gastroenteritis (AGE) in people of all age groups. In India, norovirus rates between 1.4 to 44.4% have been reported. Only a very few complete norovirus genome sequences from India have been reported.
Objective: To perform full genome sequencing of noroviruses circulating in India during 2017-2021, identify circulating genotypes, assess evolution including detection of recombination events.
Methodology: Forty-five archived norovirus-positive samples collected between October 2017 to July 2021 from patients with AGE from two hospitals in Kolkata, India were processed for full genome sequencing. Phylogenetic analysis, recombination breakpoint analysis and comprehensive mutation analysis were also performed.
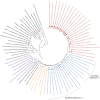
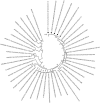
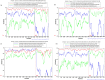
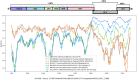
Results: Full genome analysis of norovirus sequences revealed that strains belonging to genogroup (G)I were genotyped as GI.3[P13]. Among the different norovirus capsid-polymerase combinations, GII.3[P16], GII.4 Sydney[P16], GII.4 Sydney[P31], GII.13[P16], GII.16[P16] and GII.17 were identified. Phylogenetic analysis confirmed phylogenetic relatedness with previously reported norovirus strains and all viruses were analyzed by Simplot. GII[P16] viruses with multiple residue mutations within the non-structural region were detected among circulating GII.4 and GII.3 strains. Comprehensive mutation analysis and selection pressure analysis of GII[P16] viruses showed positive as well as negative selection sites. A GII.17 strain (NICED-BCH-11889) had an untypeable polymerase type, closely related to GII[P38].
Conclusion: This study highlights the circulation of diverse norovirus strains in eastern India. These findings are important for understanding norovirus epidemiology in India and may have implications for future vaccine development.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Recombinant Noroviruses Circulating in Spain from 2016 to 2020 and Proposal of Two Novel Genotypes within Genogroup I.Microbiol Spectr. 2022 Aug 31;10(4):e0250521. doi: 10.1128/spectrum.02505-21. Epub 2022 Jul 13. Microbiol Spectr. 2022. PMID: 35862999 Free PMC article.
-
Genetic characterization and evolutionary analysis of norovirus genotypes circulating among children in eastern India during 2018-2019.Arch Virol. 2021 Nov;166(11):2989-2998. doi: 10.1007/s00705-021-05197-6. Epub 2021 Aug 12. Arch Virol. 2021. PMID: 34383167 Free PMC article.
-
Molecular epidemiology and genetic diversity of norovirus infection in children hospitalized with acute gastroenteritis in East Java, Indonesia in 2015-2019.Infect Genet Evol. 2021 Mar;88:104703. doi: 10.1016/j.meegid.2020.104703. Epub 2021 Jan 2. Infect Genet Evol. 2021. PMID: 33401005
-
Characterization of the complete genome sequence of the recombinant norovirus GII.P16/GII.4_Sydney_2012 revealed in Russia.Vavilovskii Zhurnal Genet Selektsii. 2020 Feb;24(1):69-79. doi: 10.18699/VJ20.597. Vavilovskii Zhurnal Genet Selektsii. 2020. PMID: 33659783 Free PMC article.
-
Global and regional circulation trends of norovirus genotypes and recombinants, 1995-2019: A comprehensive review of sequences from public databases.Rev Med Virol. 2022 Sep;32(5):e2354. doi: 10.1002/rmv.2354. Epub 2022 Apr 28. Rev Med Virol. 2022. PMID: 35481689 Free PMC article. Review.
Cited by
-
Conformational Flexibility in Capsids Encoded by the Caliciviridae.Viruses. 2024 Nov 26;16(12):1835. doi: 10.3390/v16121835. Viruses. 2024. PMID: 39772145 Free PMC article. Review.
-
Draft genomes of norovirus from stool samples of under-five children presenting with gastroenteritis in Malawi from 2012 to 2024.Microbiol Resour Announc. 2025 Aug 14;14(8):e0060225. doi: 10.1128/mra.00602-25. Epub 2025 Jul 28. Microbiol Resour Announc. 2025. PMID: 40719297 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

