Medicinal chemistry strategies towards the development of non-covalent SARS-CoV-2 Mpro inhibitors
- PMID: 38239241
- PMCID: PMC10792984
- DOI: 10.1016/j.apsb.2023.08.004
Medicinal chemistry strategies towards the development of non-covalent SARS-CoV-2 Mpro inhibitors
Abstract
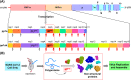
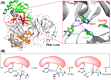
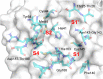
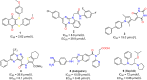
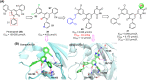
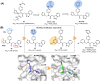
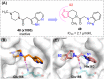
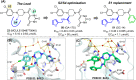
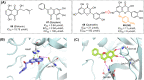
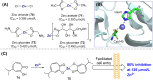
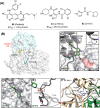
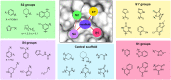
The main protease (Mpro) of SARS-CoV-2 is an attractive target in anti-COVID-19 therapy for its high conservation and major role in the virus life cycle. The covalent Mpro inhibitor nirmatrelvir (in combination with ritonavir, a pharmacokinetic enhancer) and the non-covalent inhibitor ensitrelvir have shown efficacy in clinical trials and have been approved for therapeutic use. Effective antiviral drugs are needed to fight the pandemic, while non-covalent Mpro inhibitors could be promising alternatives due to their high selectivity and favorable druggability. Numerous non-covalent Mpro inhibitors with desirable properties have been developed based on available crystal structures of Mpro. In this article, we describe medicinal chemistry strategies applied for the discovery and optimization of non-covalent Mpro inhibitors, followed by a general overview and critical analysis of the available information. Prospective viewpoints and insights into current strategies for the development of non-covalent Mpro inhibitors are also discussed.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

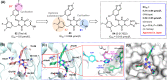

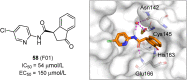
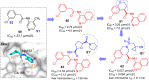
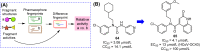

References
-
- World Health Organization. WHO coronavirus (COVID-19) dashboard. Available from: https://covid19.who.int/.
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

