Integrated analysis of single-cell RNA-seq and chipset data unravels PANoptosis-related genes in sepsis
- PMID: 38239341
- PMCID: PMC10795179
- DOI: 10.3389/fimmu.2023.1247131
Integrated analysis of single-cell RNA-seq and chipset data unravels PANoptosis-related genes in sepsis
Erratum in
-
Corrigendum: Integrated analysis of single-cell RNA-seq and chipset data unravels PANoptosis-related genes in sepsis.Front Immunol. 2024 Apr 23;15:1415915. doi: 10.3389/fimmu.2024.1415915. eCollection 2024. Front Immunol. 2024. PMID: 38715603 Free PMC article.
Abstract
Background: The poor prognosis of sepsis warrants the investigation of biomarkers for predicting the outcome. Several studies have indicated that PANoptosis exerts a critical role in tumor initiation and development. Nevertheless, the role of PANoptosis in sepsis has not been fully elucidated.
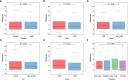
Methods: We obtained Sepsis samples and scRNA-seq data from the GEO database. PANoptosis-related genes were subjected to consensus clustering and functional enrichment analysis, followed by identification of differentially expressed genes and calculation of the PANoptosis score. A PANoptosis-based prognostic model was developed. In vitro experiments were performed to verify distinct PANoptosis-related genes. An external scRNA-seq dataset was used to verify cellular localization.
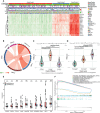
Results: Unsupervised clustering analysis using 16 PANoptosis-related genes identified three subtypes of sepsis. Kaplan-Meier analysis showed significant differences in patient survival among the subtypes, with different immune infiltration levels. Differential analysis of the subtypes identified 48 DEGs. Boruta algorithm PCA analysis identified 16 DEGs as PANoptosis-related signature genes. We developed PANscore based on these signature genes, which can distinguish different PANoptosis and clinical characteristics and may serve as a potential biomarker. Single-cell sequencing analysis identified six cell types, with high PANscore clustering relatively in B cells, and low PANscore in CD16+ and CD14+ monocytes and Megakaryocyte progenitors. ZBP1, XAF1, IFI44L, SOCS1, and PARP14 were relatively higher in cells with high PANscore.
Conclusion: We developed a machine learning based Boruta algorithm for profiling PANoptosis related subgroups with in predicting survival and clinical features in the sepsis.
Keywords: Boruta algorithm; PANoptosis; sepsis; single-cell RNA-seq; ssGSEA.
Copyright © 2024 Dai, Zheng, Wu, Chen, Deng, Tong, Liu, Shang and Qian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Anggraini D, Hasni D, Amelia R. Pathogenesis of sepsis. Sci J (2022) 1:332–9. doi: 10.56260/sciena.v1i4.63 - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

