Nutrient-dependent cross-kingdom interactions in the hyphosphere of an arbuscular mycorrhizal fungus
- PMID: 38239731
- PMCID: PMC10794670
- DOI: 10.3389/fmicb.2023.1284648
Nutrient-dependent cross-kingdom interactions in the hyphosphere of an arbuscular mycorrhizal fungus
Abstract
Introduction: The hyphosphere of arbuscular mycorrhizal (AM) fungi is teeming with microbial life. Yet, the influence of nutrient availability or nutrient forms on the hyphosphere microbiomes is still poorly understood.
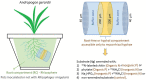
Methods: Here, we examined how the microbial community (prokaryotic, fungal, protistan) was affected by the presence of the AM fungus Rhizophagus irregularis in the rhizosphere and the root-free zone, and how different nitrogen (N) and phosphorus (P) supplements into the root-free compartment influenced the communities.
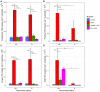
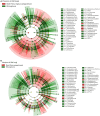
Results: The presence of AM fungus greatly affected microbial communities both in the rhizosphere and the root-free zone, with prokaryotic communities being affected the most. Protists were the only group of microbes whose richness and diversity were significantly reduced by the presence of the AM fungus. Our results showed that the type of nutrients AM fungi encounter in localized patches modulate the structure of hyphosphere microbial communities. In contrast we did not observe any effects of the AM fungus on (non-mycorrhizal) fungal community composition. Compared to the non-mycorrhizal control, the root-free zone with the AM fungus (i.e., the AM fungal hyphosphere) was enriched with Alphaproteobacteria, some micropredatory and copiotroph bacterial taxa (e.g., Xanthomonadaceae and Bacteroidota), and the poorly characterized and not yet cultured Acidobacteriota subgroup GP17, especially when phytate was added. Ammonia-oxidizing Nitrosomonas and nitrite-oxidizing Nitrospira were significantly suppressed in the presence of the AM fungus in the root-free compartment, especially upon addition of inorganic N. Co-occurrence network analyses revealed that microbial communities in the root-free compartment were complex and interconnected with more keystone species when AM fungus was present, especially when the root-free compartment was amended with phytate.
Conclusion: Our study showed that the form of nutrients is an important driver of prokaryotic and eukaryotic community assembly in the AM fungal hyphosphere, despite the assumed presence of a stable and specific AM fungal hyphoplane microbiome. Predictable responses of specific microbial taxa will open the possibility of using them as co-inoculants with AM fungi, e.g., to improve crop performance.
Keywords: arbuscular mycorrhizal (AM) fungi/al; extraradical hyphae; hyphosphere; inorganic and organic; microbiome; networks; nutrient cycling; nutrient mobilization.
Copyright © 2024 Faghihinia, Halverson, Hršelová, Bukovská, Rozmoš, Kotianová and Jansa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Bastian M., Heymann S., Jacomy M., (2009). Gephi: an open source software for exploring and manipulating networks. Proceedings of the International AAAI Conference on Weblogs and Social Media, San Jose, California, USA.
LinkOut - more resources
Full Text Sources

