The Plasma Lipidomic Landscape in Patients with Sepsis due to Community-acquired Pneumonia
- PMID: 38240721
- PMCID: PMC12039242
- DOI: 10.1164/rccm.202308-1321OC
The Plasma Lipidomic Landscape in Patients with Sepsis due to Community-acquired Pneumonia
Abstract
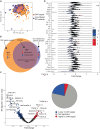
Rationale: The plasma lipidome has the potential to reflect many facets of the host status during severe infection. Previous work is limited to specific lipid groups or was focused on lipids as prognosticators.Objectives: To map the plasma lipidome during sepsis due to community-acquired pneumonia (CAP) and determine the disease specificity and associations with clinical features.Methods: We analyzed 1,833 lipid species across 33 classes in 169 patients admitted to the ICU with sepsis due to CAP, 51 noninfected ICU patients, and 48 outpatient controls. In a paired analysis, we reanalyzed patients still in the ICU 4 days after admission (n = 82).Measurements and Main Results: A total of 58% of plasma lipids were significantly lower in patients with CAP-attributable sepsis compared with outpatient controls (6% higher, 36% not different). We found strong lipid class-specific associations with disease severity, validated across two external cohorts, and inflammatory biomarkers, in which triacylglycerols, cholesterol esters, and lysophospholipids exhibited the strongest associations. A total of 36% of lipids increased over time, and stratification by survival revealed diverging lipid recovery, which was confirmed in an external cohort; specifically, a 10% increase in cholesterol ester levels was related to a lower odds ratio (0.84; P = 0.006) for 30-day mortality (absolute mortality, 18 of 82). Comparison with noninfected ICU patients delineated a substantial common illness response (57.5%) and a distinct lipidomic signal for patients with CAP-attributable sepsis (37%).Conclusions: Patients with sepsis due to CAP exhibit a time-dependent and partially disease-specific shift in their plasma lipidome that correlates with disease severity and systemic inflammation and is associated with higher mortality.
Keywords: cholesterol; lipidomics; respiratory failure; sepsis; triglycerides.
Figures
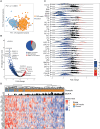

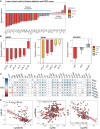
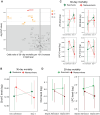
Comment in
-
A Lipid Map for Community-acquired Pneumonia with Sepsis: Observation Is the First Step in Scientific Progress.Am J Respir Crit Care Med. 2024 Apr 15;209(8):903-904. doi: 10.1164/rccm.202401-0213ED. Am J Respir Crit Care Med. 2024. PMID: 38412325 Free PMC article. No abstract available.
References
-
- Torres A, Cilloniz C, Niederman MS, Menéndez R, Chalmers JD, Wunderink RG, et al. Pneumonia. Nat Rev Dis Primers . 2021;7:25. - PubMed
-
- van der Poll T, Shankar-Hari M, Wiersinga WJ. The immunology of sepsis. Immunity . 2021;54:2450–2464. - PubMed
-
- Scicluna BP, van Vught LA, Zwinderman AH, Wiewel MA, Davenport EE, Burnham KL, et al. MARS consortium Classification of patients with sepsis according to blood genomic endotype: a prospective cohort study. Lancet Respir Med . 2017;5:816–826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

