Mechanism of exportin retention in the cell nucleus
- PMID: 38241019
- PMCID: PMC10798875
- DOI: 10.1083/jcb.202306094
Mechanism of exportin retention in the cell nucleus
Abstract
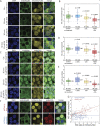
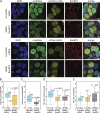
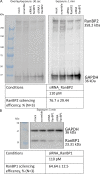
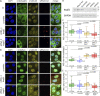
Exportin receptors are concentrated in the nucleus to transport essential cargoes out of it. A mislocalization of exportins to the cytoplasm is linked to disease. Hence, it is important to understand how their containment within the nucleus is regulated. Here, we have studied the nuclear efflux of exportin2 (cellular apoptosis susceptibility protein or CAS) that delivers karyopherinα (Kapα or importinα), the cargo adaptor for karyopherinβ1 (Kapβ1 or importinβ1), to the cytoplasm in a Ran guanosine triphosphate (RanGTP)-mediated manner. We show that the N-terminus of CAS attenuates the interaction of RanGTPase activating protein 1 (RanGAP1) with RanGTP to slow GTP hydrolysis, which suppresses CAS nuclear exit at nuclear pore complexes (NPCs). Strikingly, a single phosphomimetic mutation (T18D) at the CAS N-terminus is sufficient to abolish its nuclear retention and coincides with metastatic cellular behavior. Furthermore, downregulating Kapβ1 disrupts CAS nuclear retention, which highlights the balance between their respective functions that is essential for maintaining the Kapα transport cycle. Therefore, NPCs play a functional role in selectively partitioning exportins in the cell nucleus.
© 2024 Kapinos et al.
Conflict of interest statement
Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. L.E. Kapinos reported “This work is funded by the Swiss National Science Foundation project no. 310030_201062.” R.Y.H. Lim reported “This work is funded by the Swiss National Science Foundation project no. 310030_201062.” No other disclosures were reported.
Figures








Similar articles
-
Karyopherin enrichment at the nuclear pore complex attenuates Ran permeability.J Cell Sci. 2020 Feb 12;133(3):jcs238121. doi: 10.1242/jcs.238121. J Cell Sci. 2020. PMID: 31932502
-
Ran-dependent nuclear export mediators: a structural perspective.EMBO J. 2011 Aug 31;30(17):3457-74. doi: 10.1038/emboj.2011.287. EMBO J. 2011. PMID: 21878989 Free PMC article. Review.
-
Karyopherins regulate nuclear pore complex barrier and transport function.J Cell Biol. 2017 Nov 6;216(11):3609-3624. doi: 10.1083/jcb.201702092. Epub 2017 Sep 1. J Cell Biol. 2017. PMID: 28864541 Free PMC article.
-
On the asymmetric partitioning of nucleocytoplasmic transport - recent insights and open questions.J Cell Sci. 2021 Apr 1;134(7):jcs240382. doi: 10.1242/jcs.240382. Epub 2021 Apr 13. J Cell Sci. 2021. PMID: 33912945 Review.
-
Mechanistic Insights from Structural Analyses of Ran-GTPase-Driven Nuclear Export of Proteins and RNAs.J Mol Biol. 2016 May 22;428(10 Pt A):2025-39. doi: 10.1016/j.jmb.2015.09.025. Epub 2015 Oct 28. J Mol Biol. 2016. PMID: 26519791 Review.
Cited by
-
Elucidating the nanoscopic organization and dynamics of the nuclear pore complex.Nucleus. 2025 Dec;16(1):2510106. doi: 10.1080/19491034.2025.2510106. Epub 2025 Jun 4. Nucleus. 2025. PMID: 40464641 Free PMC article.
-
Strategies for the Viral Exploitation of Nuclear Pore Transport Pathways.Viruses. 2025 Jan 23;17(2):151. doi: 10.3390/v17020151. Viruses. 2025. PMID: 40006906 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

