Evaluating deep learning-based melanoma classification using immunohistochemistry and routine histology: A three center study
- PMID: 38241314
- PMCID: PMC10798511
- DOI: 10.1371/journal.pone.0297146
Evaluating deep learning-based melanoma classification using immunohistochemistry and routine histology: A three center study
Abstract
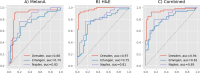
Pathologists routinely use immunohistochemical (IHC)-stained tissue slides against MelanA in addition to hematoxylin and eosin (H&E)-stained slides to improve their accuracy in diagnosing melanomas. The use of diagnostic Deep Learning (DL)-based support systems for automated examination of tissue morphology and cellular composition has been well studied in standard H&E-stained tissue slides. In contrast, there are few studies that analyze IHC slides using DL. Therefore, we investigated the separate and joint performance of ResNets trained on MelanA and corresponding H&E-stained slides. The MelanA classifier achieved an area under receiver operating characteristics curve (AUROC) of 0.82 and 0.74 on out of distribution (OOD)-datasets, similar to the H&E-based benchmark classification of 0.81 and 0.75, respectively. A combined classifier using MelanA and H&E achieved AUROCs of 0.85 and 0.81 on the OOD datasets. DL MelanA-based assistance systems show the same performance as the benchmark H&E classification and may be improved by multi stain classification to assist pathologists in their clinical routine.
Copyright: © 2024 Wies et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
TJB would like to disclose that he is the owner of Smart Health Heidelberg GmbH (Handschuhsheimer Landstr. 9/1, 69120 Heidelberg, Germany) which develops mobile apps, outside of the submitted work. SHo reports clinical trial support from Almirall and speaker’s honoraria from Almirall, UCB and AbbVie and has received travel support from the following companies: UCB, Janssen Cilag, Almirall, Novartis, Lilly, LEO Pharma and AbbVie outside the submitted work. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures
Similar articles
-
Deep learning-based subtyping of gastric cancer histology predicts clinical outcome: a multi-institutional retrospective study.Gastric Cancer. 2023 Sep;26(5):708-720. doi: 10.1007/s10120-023-01398-x. Epub 2023 Jun 3. Gastric Cancer. 2023. PMID: 37269416 Free PMC article.
-
Piloting a Deep Learning Model for Predicting Nuclear BAP1 Immunohistochemical Expression of Uveal Melanoma from Hematoxylin-and-Eosin Sections.Transl Vis Sci Technol. 2020 Sep 1;9(2):50. doi: 10.1167/tvst.9.2.50. eCollection 2020 Sep. Transl Vis Sci Technol. 2020. PMID: 32953248 Free PMC article.
-
Deep learning detection of melanoma metastases in lymph nodes.Eur J Cancer. 2023 Jul;188:161-170. doi: 10.1016/j.ejca.2023.04.023. Epub 2023 Apr 29. Eur J Cancer. 2023. PMID: 37257277
-
Histopathologic review of negative sentinel lymph node biopsies in thin melanomas: an argument for the routine use of immunohistochemistry.Melanoma Res. 2017 Aug;27(4):369-376. doi: 10.1097/CMR.0000000000000361. Melanoma Res. 2017. PMID: 28654548 Review.
-
Utilization of immunohistochemistry in gynecologic tumors: An expert review.Gynecol Oncol Rep. 2024 Nov 29;56:101550. doi: 10.1016/j.gore.2024.101550. eCollection 2024 Dec. Gynecol Oncol Rep. 2024. PMID: 39717157 Free PMC article. Review.
Cited by
-
Automated Detection of Kaposi Sarcoma-Associated Herpesvirus-Infected Cells in Immunohistochemical Images of Skin Biopsies.JCO Glob Oncol. 2025 Apr;11:e2400536. doi: 10.1200/GO-24-00536. Epub 2025 Apr 16. JCO Glob Oncol. 2025. PMID: 40239145
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

