Engineered cocultures of iPSC-derived atrial cardiomyocytes and atrial fibroblasts for modeling atrial fibrillation
- PMID: 38241367
- PMCID: PMC10798559
- DOI: 10.1126/sciadv.adg1222
Engineered cocultures of iPSC-derived atrial cardiomyocytes and atrial fibroblasts for modeling atrial fibrillation
Abstract
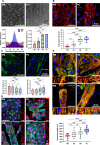
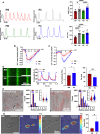
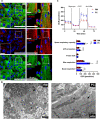
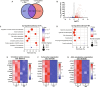
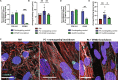
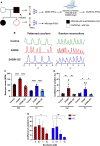
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia treatable with antiarrhythmic drugs; however, patient responses remain highly variable. Human induced pluripotent stem cell-derived atrial cardiomyocytes (iPSC-aCMs) are useful for discovering precision therapeutics, but current platforms yield phenotypically immature cells and are not easily scalable for high-throughput screening. Here, primary adult atrial, but not ventricular, fibroblasts induced greater functional iPSC-aCM maturation, partly through connexin-40 and ephrin-B1 signaling. We developed a protein patterning process within multiwell plates to engineer patterned iPSC-aCM and atrial fibroblast coculture (PC) that significantly enhanced iPSC-aCM structural, electrical, contractile, and metabolic maturation for 6+ weeks compared to conventional mono-/coculture. PC displayed greater sensitivity for detecting drug efficacy than monoculture and enabled the modeling and pharmacological or gene editing treatment of an AF-like electrophysiological phenotype due to a mutated sodium channel. Overall, PC is useful for elucidating cell signaling in the atria, drug screening, and modeling AF.
Figures


References
-
- Chugh S. S., Havmoeller R., Narayanan K., Singh D., Rienstra M., Benjamin E. J., Gillum R. F., Kim Y. H., McAnulty J. H. Jr., Zheng Z. J., Forouzanfar M. H., Naghavi M., Mensah G. A., Ezzati M., Murray C. J. L., Worldwide epidemiology of atrial fibrillation: A global burden of Disease 2010 Study. Circulation 129, 837–847 (2014). - PMC - PubMed
-
- Wijesurendra R. S., Casadei B., Mechanisms of atrial fibrillation. Heart 105, 1860–1867 (2019). - PubMed
-
- Chen Y. H., Xu S. J., Bendahhou S., Wang X. L., Wang Y., Xu W. Y., Jin H. W., Sun H., Su X. Y., Zhuang Q. N., Yang Y. Q., Li Y. B., Liu Y., Xu H. J., Li X. F., Ma N., Mou C. P., Chen Z., Barhanin J., Huang W., KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science 299, 251–254 (2003). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

