Inhibiting the Keap1/Nrf2 Protein-Protein Interaction with Protein-Like Polymers
- PMID: 38241649
- PMCID: PMC11257647
- DOI: 10.1002/adma.202311467
Inhibiting the Keap1/Nrf2 Protein-Protein Interaction with Protein-Like Polymers
Abstract
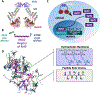
Successful and selective inhibition of the cytosolic protein-protein interaction (PPI) between nuclear factor erythroid 2-related factor 2 (Nrf2) and Kelch-like ECH-associating protein 1 (Keap1) can enhance the antioxidant response, with the potential for a therapeutic effect in a range of settings including in neurodegenerative disease (ND). Small molecule inhibitors have been developed, yet many have off-target effects, or are otherwise limited by poor cellular permeability. Peptide-based strategies have also been attempted to enhance specificity, yet face challenges due to susceptibility to degradation and lack of cellular penetration. Herein, these barriers are overcome utilizing a polymer-based proteomimetics. The protein-like polymer (PLP) consists of a synthetic, lipophilic polymer backbone displaying water soluble Keap1-binding peptides on each monomer unit forming a brush polymer architecture. The PLPs are capable of engaging Keap1 and displacing the cellular protective transcription factor Nrf2, which then translocates to the nucleus, activating the antioxidant response element (ARE). PLPs exhibit increased Keap1 binding affinity by several orders of magnitude compared to free peptides, maintain serum stability, are cell-penetrant, and selectively activate the ARE pathway in cells, including in primary cortical neuronal cultures. Keap1/Nrf2-inhibitory PLPs have the potential to impact the treatment of disease states associated with dysregulation of oxidative stress, such as NDs.
Keywords: antioxidant; biomaterial; drug delivery; peptides; polymers.
© 2024 The Authors. Advanced Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
Competing Interests:
N.C.G and P.A.B are co-Founders of Grove Biopharma, which is a licensee of Intellectual Property (IP) related to the science and materials found in this manuscript. N.C.G and K.P.C. are co-inventors on that same IP. J.A.J and N.C.G serve on the Scientific Advisory Board for Grove Biopharma.
Figures


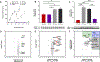

References
MeSH terms
Substances
Grants and funding
- CHE-1048773/National Science Foundation
- R00CA248715/NH/NIH HHS/United States
- DGE1745303/National Science Foundation
- 2R01HL139001-05A1/NH/NIH HHS/United States
- R01 HL139001/HL/NHLBI NIH HHS/United States
- R00 CA248715/CA/NCI NIH HHS/United States
- P30CA060553/CA/NCI NIH HHS/United States
- RyanFellowship/International Institute for Nanotechnology, Northwestern University
- T32 GM149439/GM/NIGMS NIH HHS/United States
- T32GM149439/NH/NIH HHS/United States
- F30 AG076317/AG/NIA NIH HHS/United States
- DFS-53-22/DRCRF/Damon Runyon Cancer Research Foundation/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- 1F30AG076317-01A1/AG/NIA NIH HHS/United States
- DMR-2004899/National Science Foundation
- Sherman and Fairchild Foundation
- 1F31AG076334-01/NH/NIH HHS/United States
- ECCS-2025633/National Science Foundation
LinkOut - more resources
Full Text Sources
Research Materials

