AKT1 interacts with DHX9 to Mitigate R Loop-Induced Replication Stress in Ovarian Cancer
- PMID: 38241710
- PMCID: PMC10947874
- DOI: 10.1158/0008-5472.CAN-23-1908
AKT1 interacts with DHX9 to Mitigate R Loop-Induced Replication Stress in Ovarian Cancer
Abstract
PARP inhibitor (PARPi)-resistant BRCA-mutant (BRCAm) high-grade serous ovarian cancer (HGSOC) represents a new clinical challenge with unmet therapeutic needs. Here, we performed a quantitative high-throughput drug combination screen that identified the combination of an ATR inhibitor (ATRi) and an AKT inhibitor (AKTi) as an effective treatment strategy for both PARPi-sensitive and PARPi-resistant BRCAm HGSOC. The ATRi and AKTi combination induced DNA damage and R loop-mediated replication stress (RS). Mechanistically, the kinase domain of AKT1 directly interacted with DHX9 and facilitated recruitment of DHX9 to R loops. AKTi increased ATRi-induced R loop-mediated RS by mitigating recruitment of DHX9 to R loops. Moreover, DHX9 was upregulated in tumors from patients with PARPi-resistant BRCAm HGSOC, and high coexpression of DHX9 and AKT1 correlated with worse survival. Together, this study reveals an interaction between AKT1 and DHX9 that facilitates R loop resolution and identifies combining ATRi and AKTi as a rational treatment strategy for BRCAm HGSOC irrespective of PARPi resistance status.
Significance: Inhibition of the AKT and ATR pathways cooperatively induces R loop-associated replication stress in high-grade serous ovarian cancer, providing rationale to support the clinical development of AKT and ATR inhibitor combinations. See related commentary by Ramanarayanan and Oberdoerffer, p. 793.
©2024 American Association for Cancer Research.
Conflict of interest statement
Figures


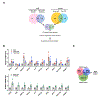
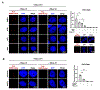
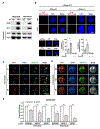


Comment in
-
AKTing on R Loops Makes for an ATRactive Target in Ovarian Cancer Therapy.Cancer Res. 2024 Mar 15;84(6):793-795. doi: 10.1158/0008-5472.CAN-23-4129. Cancer Res. 2024. PMID: 38486481
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209–49 - PubMed
-
- Armstrong DK, Alvarez RD, Backes FJ, Bakkum-Gamez JN, Barroilhet L, Behbakht K, et al. NCCN Guidelines(R) Insights: Ovarian Cancer, Version 3.2022. J Natl Compr Canc Netw 2022;20:972–80 - PubMed
-
- Dias MP, Moser SC, Ganesan S, Jonkers J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat Rev Clin Oncol 2021;18:773–91 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

