Surface CD52, CD84, and PTGER2 mark mature PMN-MDSCs from cancer patients and G-CSF-treated donors
- PMID: 38242120
- PMCID: PMC10897522
- DOI: 10.1016/j.xcrm.2023.101380
Surface CD52, CD84, and PTGER2 mark mature PMN-MDSCs from cancer patients and G-CSF-treated donors
Abstract
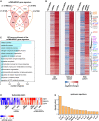
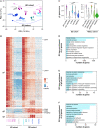
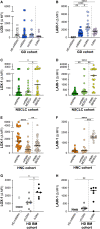
Precise molecular characterization of circulating polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) is hampered by their mixed composition of mature and immature cells and lack of specific markers. Here, we focus on mature CD66b+CD10+CD16+CD11b+ PMN-MDSCs (mPMN-MDSCs) from either cancer patients or healthy donors receiving G-CSF for stem cell mobilization (GDs). By RNA sequencing (RNA-seq) experiments, we report the identification of a distinct gene signature shared by the different mPMN-MDSC populations under investigation, also validated in mPMN-MDSCs from GDs and tumor-associated neutrophils (TANs) by single-cell RNA-seq (scRNA-seq) experiments. Analysis of such a gene signature uncovers a specific transcriptional program associated with mPMN-MDSC differentiation and allows us to identify that, in patients with either solid or hematologic tumors and in GDs, CD52, CD84, and prostaglandin E receptor 2 (PTGER2) represent potential mPMN-MDSC-associated markers. Altogether, our findings indicate that mature PMN-MDSCs distinctively undergo specific reprogramming during differentiation and lay the groundwork for selective immunomonitoring, and eventually targeting, of mature PMN-MDSCs.
Keywords: G-CSF; PMN-MDSCs; biomarkers; cancer patients; neutrophils.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Hacbarth E., Kajdacsy-Balla A. Low density neutrophils in patients with systemic lupus erythematosus, rheumatoid arthritis, and acute rheumatic fever. Arthritis Rheum. 1986;29:1334–1342. - PubMed
-
- Scapini P., Marini O., Tecchio C., Cassatella M.A. Human neutrophils in the saga of cellular heterogeneity: insights and open questions. Immunol. Rev. 2016;273:48–60. - PubMed
-
- Denny M.F., Yalavarthi S., Zhao W., Thacker S.G., Anderson M., Sandy A.R., McCune W.J., Kaplan M.J. A distinct subset of proinflammatory neutrophils isolated from patients with systemic lupus erythematosus induces vascular damage and synthesizes type I IFNs. J. Immunol. 2010;184:3284–3297. - PMC - PubMed
-
- Ochoa A.C., Zea A.H., Hernandez C., Rodriguez P.C. Arginase, prostaglandins, and myeloid-derived suppressor cells in renal cell carcinoma. Clin. Cancer Res. 2007;13:721s–726s. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

