Elevated cholesterol in ATAD3 mutants is a compensatory mechanism that leads to membrane cholesterol aggregation
- PMID: 38242545
- PMCID: PMC11068212
- DOI: 10.1093/brain/awae018
Elevated cholesterol in ATAD3 mutants is a compensatory mechanism that leads to membrane cholesterol aggregation
Abstract
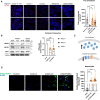
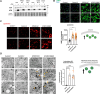
Aberrant cholesterol metabolism causes neurological disease and neurodegeneration, and mitochondria have been linked to perturbed cholesterol homeostasis via the study of pathological mutations in the ATAD3 gene cluster. However, whether the cholesterol changes were compensatory or contributory to the disorder was unclear, and the effects on cell membranes and the wider cell were also unknown. Using patient-derived cells, we show that cholesterol perturbation is a conserved feature of pathological ATAD3 variants that is accompanied by an expanded lysosome population containing membrane whorls characteristic of lysosomal storage diseases. Lysosomes are also more numerous in Drosophila neural progenitor cells expressing mutant Atad3, which exhibit abundant membrane-bound cholesterol aggregates, many of which co-localize with lysosomes. By subjecting the Drosophila Atad3 mutant to nutrient restriction and cholesterol supplementation, we show that the mutant displays heightened cholesterol dependence. Collectively, these findings suggest that elevated cholesterol enhances tolerance to pathological ATAD3 variants; however, this comes at the cost of inducing cholesterol aggregation in membranes, which lysosomal clearance only partly mitigates.
Keywords: AAA+ ATPase; ATAD3; cholesterol disorders; lysosomal storage disorders; lysosomes; mitochondrial disease.
© The Author(s) 2024. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
The authors report no competing interests.
Figures





References
-
- Pfrieger FW. The Niemann-Pick type diseases—A synopsis of inborn errors in sphingolipid and cholesterol metabolism. Prog Lipid Res. 2023;90:101225. - PubMed
-
- Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol. 2020;21:225–245. - PubMed
-
- Shamitko-Klingensmith N, Molchanoff KM, Burke KA, Magnone GJ, Legleiter J. Mapping the mechanical properties of cholesterol-containing supported lipid bilayers with nanoscale spatial resolution. Langmuir. 2012;28:13411–13422. - PubMed
-
- Borochov H, Abbott RE, Schachter D, Shinitzky M. Modulation of erythrocyte membrane proteins by membrane cholesterol and lipid fluidity. Biochemistry. 1979;18:251–255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

