Improving prenatal diagnosis through standards and aggregation
- PMID: 38242839
- PMCID: PMC11006584
- DOI: 10.1002/pd.6522
Improving prenatal diagnosis through standards and aggregation
Abstract
Advances in sequencing and imaging technologies enable enhanced assessment in the prenatal space, with a goal to diagnose and predict the natural history of disease, to direct targeted therapies, and to implement clinical management, including transfer of care, election of supportive care, and selection of surgical interventions. The current lack of standardization and aggregation stymies variant interpretation and gene discovery, which hinders the provision of prenatal precision medicine, leaving clinicians and patients without an accurate diagnosis. With large amounts of data generated, it is imperative to establish standards for data collection, processing, and aggregation. Aggregated and homogeneously processed genetic and phenotypic data permits dissection of the genomic architecture of prenatal presentations of disease and provides a dataset on which data analysis algorithms can be tuned to the prenatal space. Here we discuss the importance of generating aggregate data sets and how the prenatal space is driving the development of interoperable standards and phenotype-driven tools.
© 2024 The Authors. Prenatal Diagnosis published by John Wiley & Sons Ltd.
Conflict of interest statement
M.E.T. and H.B. receive research funding from Microsoft Inc. R.J.W., M.E.T. and H.B. received reagents from Illumina Inc.
Figures

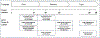


References
Publication types
MeSH terms
Grants and funding
- R01 HD103805/HD/NICHD NIH HHS/United States
- 1R01HD103805/NH/NIH HHS/United States
- R24 OD011883/OD/NIH HHS/United States
- UM1 HG006370/HG/NHGRI NIH HHS/United States
- 1F32HD112084/NH/NIH HHS/United States
- U54 HG006370/HG/NHGRI NIH HHS/United States
- RM1 HG010860/HG/NHGRI NIH HHS/United States
- 5RM1HG010860/NH/NIH HHS/United States
- 5R01HD105266/NH/NIH HHS/United States
- 5R24OD011883/NH/NIH HHS/United States
- UM1HG006370/NH/NIH HHS/United States
- R01 HD055651/HD/NICHD NIH HHS/United States
- R01 HD081256/HD/NICHD NIH HHS/United States
- 5R01HD055651/NH/NIH HHS/United States
- U24 HG011449/HG/NHGRI NIH HHS/United States
- R01 HD105266/HD/NICHD NIH HHS/United States
- 3R01HD055651-15S1/NH/NIH HHS/United States
- F32 HD112084/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

