Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations
- PMID: 38243071
- PMCID: PMC10799026
- DOI: 10.1038/s42003-023-05726-w
Differences in regulation mechanisms of glutamine synthetases from methanogenic archaea unveiled by structural investigations
Abstract
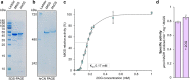
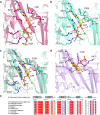
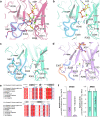
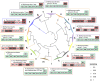
Glutamine synthetases (GS) catalyze the ATP-dependent ammonium assimilation, the initial step of nitrogen acquisition that must be under tight control to fit cellular needs. While their catalytic mechanisms and regulations are well-characterized in bacteria and eukaryotes, only limited knowledge exists in archaea. Here, we solved two archaeal GS structures and unveiled unexpected differences in their regulatory mechanisms. GS from Methanothermococcus thermolithotrophicus is inactive in its resting state and switched on by 2-oxoglutarate, a sensor of cellular nitrogen deficiency. The enzyme activation overlays remarkably well with the reported cellular concentration for 2-oxoglutarate. Its binding to an allosteric pocket reconfigures the active site through long-range conformational changes. The homolog from Methermicoccus shengliensis does not harbor the 2-oxoglutarate binding motif and, consequently, is 2-oxoglutarate insensitive. Instead, it is directly feedback-inhibited through glutamine recognition by the catalytic Asp50'-loop, a mechanism common to bacterial homologs, but absent in M. thermolithotrophicus due to residue substitution. Analyses of residue conservation in archaeal GS suggest that both regulations are widespread and not mutually exclusive. While the effectors and their binding sites are surprisingly different, the molecular mechanisms underlying their mode of action on GS activity operate on the same molecular determinants in the active site.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
2-oxoglutarate triggers assembly of active dodecameric Methanosarcina mazei glutamine synthetase.Elife. 2025 Mar 31;13:RP97484. doi: 10.7554/eLife.97484. Elife. 2025. PMID: 40163028 Free PMC article.
-
Glutamine Synthetase: Diverse Regulation and Functions of an Ancient Enzyme.Biochemistry. 2025 Feb 4;64(3):547-554. doi: 10.1021/acs.biochem.4c00763. Epub 2025 Jan 22. Biochemistry. 2025. PMID: 39844577 Free PMC article. Review.
-
M. mazei glutamine synthetase and glutamine synthetase-GlnK1 structures reveal enzyme regulation by oligomer modulation.Nat Commun. 2023 Nov 15;14(1):7375. doi: 10.1038/s41467-023-43243-w. Nat Commun. 2023. PMID: 37968329 Free PMC article.
-
Molecular dissection of the glutamine synthetase-GlnR nitrogen regulatory circuitry in Gram-positive bacteria.Nat Commun. 2022 Jul 1;13(1):3793. doi: 10.1038/s41467-022-31573-0. Nat Commun. 2022. PMID: 35778410 Free PMC article.
-
Nitrogen regulation in bacteria and archaea.Annu Rev Microbiol. 2007;61:349-77. doi: 10.1146/annurev.micro.61.080706.093409. Annu Rev Microbiol. 2007. PMID: 17506680 Review.
Cited by
-
Regulation of the Ald gene encoding alanine dehydrogenase and its induction of ammonium-tolerant nitrogen fixation in Paenibacillus polymyxa WLY78.Microb Cell Fact. 2025 Aug 21;24(1):193. doi: 10.1186/s12934-025-02823-9. Microb Cell Fact. 2025. PMID: 40841905 Free PMC article.
-
2-oxoglutarate triggers assembly of active dodecameric Methanosarcina mazei glutamine synthetase.Elife. 2025 Mar 31;13:RP97484. doi: 10.7554/eLife.97484. Elife. 2025. PMID: 40163028 Free PMC article.
-
Strategies of Environmental Adaptation in the Haloarchaeal Genera Haloarcula and Natrinema.Microorganisms. 2025 Mar 27;13(4):761. doi: 10.3390/microorganisms13040761. Microorganisms. 2025. PMID: 40284598 Free PMC article.
-
Glutamine Synthetase: Diverse Regulation and Functions of an Ancient Enzyme.Biochemistry. 2025 Feb 4;64(3):547-554. doi: 10.1021/acs.biochem.4c00763. Epub 2025 Jan 22. Biochemistry. 2025. PMID: 39844577 Free PMC article. Review.
-
Evaluation of nitrogen fixation in the marine purple photosynthetic bacterium Rhodovulum sulfidophilum under autotrophic and heterotrophic conditions.Sci Rep. 2025 May 26;15(1):18344. doi: 10.1038/s41598-025-03605-4. Sci Rep. 2025. PMID: 40419626 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

