FFAR4 activation inhibits lung adenocarcinoma via blocking respiratory chain complex assembly associated mitochondrial metabolism
- PMID: 38243188
- PMCID: PMC10799372
- DOI: 10.1186/s11658-024-00535-3
FFAR4 activation inhibits lung adenocarcinoma via blocking respiratory chain complex assembly associated mitochondrial metabolism
Abstract
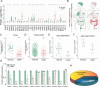
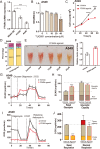
Despite notable advancements in the investigation and management of lung adenocarcinoma (LUAD), the mortality rate for individuals afflicted with LUAD remains elevated, and attaining an accurate prognosis is challenging. LUAD exhibits intricate genetic and environmental components, and it is plausible that free fatty acid receptors (FFARs) may bridge the genetic and dietary aspects. The objective of this study is to ascertain whether a correlation exists between FFAR4, which functions as the primary receptor for dietary fatty acids, and various characteristics of LUAD, while also delving into the potential underlying mechanism. The findings of this study indicate a decrease in FFAR4 expression in LUAD, with a positive correlation (P < 0.01) between FFAR4 levels and overall patient survival (OS). Receiver operating characteristic (ROC) curve analysis demonstrated a significant diagnostic value [area under the curve (AUC) of 0.933] associated with FFAR4 expression. Functional investigations revealed that the FFAR4-specific agonist (TUG891) effectively suppressed cell proliferation and induced cell cycle arrest. Furthermore, FFAR4 activation resulted in significant metabolic shifts, including a decrease in oxygen consumption rate (OCR) and an increase in extracellular acidification rate (ECAR) in A549 cells. In detail, the activation of FFAR4 has been observed to impact the assembly process of the mitochondrial respiratory chain complex and the malate-aspartate shuttle process, resulting in a decrease in the transition of NAD+ to NADH and the inhibition of LUAD. These discoveries reveal a previously unrecognized function of FFAR4 in the negative regulation of mitochondrial metabolism and the inhibition of LUAD, indicating its potential as a promising therapeutic target for the treatment and diagnosis of LUAD.
Keywords: FFAR4; LUAD; Metabolism reprogramming; OXPHOS.
© 2024. The Author(s).
Conflict of interest statement
The authors have no competing interests to declare.
Figures




Similar articles
-
High Expression of COA6 Is Related to Unfavorable Prognosis and Enhanced Oxidative Phosphorylation in Lung Adenocarcinoma.Int J Mol Sci. 2023 Mar 16;24(6):5705. doi: 10.3390/ijms24065705. Int J Mol Sci. 2023. PMID: 36982777 Free PMC article.
-
KIAA1429 promotes the progression of lung adenocarcinoma by regulating the m6A level of MUC3A.Pathol Res Pract. 2021 Jan;217:153284. doi: 10.1016/j.prp.2020.153284. Epub 2020 Nov 12. Pathol Res Pract. 2021. PMID: 33249400
-
A Novel Role of Arrhythmia-Related Gene KCNQ1 Revealed by Multi-Omic Analysis: Theragnostic Value and Potential Mechanisms in Lung Adenocarcinoma.Int J Mol Sci. 2022 Feb 18;23(4):2279. doi: 10.3390/ijms23042279. Int J Mol Sci. 2022. PMID: 35216393 Free PMC article.
-
MTHFD2 promotes tumorigenesis and metastasis in lung adenocarcinoma by regulating AKT/GSK-3β/β-catenin signalling.J Cell Mol Med. 2021 Jul;25(14):7013-7027. doi: 10.1111/jcmm.16715. Epub 2021 Jun 13. J Cell Mol Med. 2021. PMID: 34121323 Free PMC article.
-
FAM64A: A Novel Oncogenic Target of Lung Adenocarcinoma Regulated by Both Strands of miR-99a (miR-99a-5p and miR-99a-3p).Cells. 2020 Sep 11;9(9):2083. doi: 10.3390/cells9092083. Cells. 2020. PMID: 32932948 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

