Non-canonical antigens are the largest fraction of peptides presented by MHC class I in mismatch repair deficient murine colorectal cancer
- PMID: 38243308
- PMCID: PMC10797964
- DOI: 10.1186/s13073-023-01275-3
Non-canonical antigens are the largest fraction of peptides presented by MHC class I in mismatch repair deficient murine colorectal cancer
Abstract
Background: Immunotherapy based on checkpoint inhibitors is highly effective in mismatch repair deficient (MMRd) colorectal cancer (CRC). These tumors carry a high number of mutations, which are predicted to translate into a wide array of neoepitopes; however, a systematic classification of the neoantigen repertoire in MMRd CRC is lacking. Mass spectrometry peptidomics has demonstrated the existence of MHC class I associated peptides (MAPs) originating from non-coding DNA regions. Based on these premises we investigated DNA genomic regions responsible for generating MMRd-induced peptides.
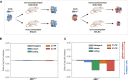
Methods: We exploited mouse CRC models in which the MMR gene Mlh1 was genetically inactivated. Isogenic cell lines CT26 Mlh1+/+ and Mlh1-/- were inoculated in immunocompromised and immunocompetent mice. Whole genome and RNA sequencing data were generated from samples obtained before and after injection in murine hosts. First, peptide databases were built from transcriptomes of isogenic cell lines. We then compiled a database of peptides lost after tumor cells injection in immunocompetent mice, likely due to immune editing. Liquid chromatography-tandem mass spectrometry (LC-MS/MS) and matched next-generation sequencing databases were employed to identify the DNA regions from which the immune-targeted MAPs originated. Finally, we adopted in vitro T cell assays to verify whether MAP-specific T cells were part of the in vivo immune response against Mlh1-/- cells.
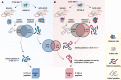
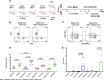
Results: Whole genome sequencing analyses revealed an unbalanced distribution of immune edited alterations across the genome in Mlh1-/- cells grown in immunocompetent mice. Specifically, untranslated (UTR) and coding regions exhibited the largest fraction of mutations leading to highly immunogenic peptides. Moreover, the integrated computational and LC-MS/MS analyses revealed that MAPs originate mainly from atypical translational events in both Mlh1+/+ and Mlh1-/- tumor cells. In addition, mutated MAPs-derived from UTRs and out-of-frame translation of coding regions-were highly enriched in Mlh1-/- cells. The MAPs trigger T-cell activation in mice primed with Mlh1-/- cells.
Conclusions: Our results suggest that-in comparison to MMR proficient CRC-MMRd tumors generate a significantly higher number of non-canonical mutated peptides able to elicit T cell responses. These results reveal the importance of evaluating the diversity of neoepitope repertoire in MMRd tumors.
Keywords: Colorectal cancer; HLA-peptidomics; Immune surveillance; MAPs; Mismatch repair; Neoantigens; Next-generation sequencing; Non-canonical antigens; Non-coding DNA.
© 2024. The Author(s).
Conflict of interest statement
A. Bardelli served in a consulting/advisory role for Illumina and Inivata. A. Bardelli and G.G. are cofounders and shareholders of NeoPhore. A. Bardelli is a member of the NeoPhore scientific advisory board. The remaining authors declare that they do not have any competing interests.
Figures


References
-
- Germano G, Amirouchene-Angelozzi N, Rospo G, Bardelli A. The clinical impact of the genomic landscape of mismatch repair-deficient cancers. Cancer Discov. 2018;8(12):1518–1528. doi: 10.1158/2159-8290.CD-18-0150. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- ID. 21091 program/Fondazione AIRC under 5 per Mille 2018
- Grant-ID 24604/AIRC MFAG 2020
- ID. 21923 project/AIRC under IG 2018
- Cancer Research UK (A26825 and A28223), FC AECC (GEACC18004TAB), and AIRC (22795)/International Accelerator Award, ACRCelerate
- grant agreement no.101020342/European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program
- contract n. 101007937/IMI PERSIST-SEQ
- 21763/AIRC under IG 2018
- 22737/AIRC 5x1000
- 101020342/ERC_/European Research Council/International
- A26825/CRUK_/Cancer Research UK/United Kingdom
- A28223/CRUK_/Cancer Research UK/United Kingdom
- GEACC18004TAB/CRUK_/Cancer Research UK/United Kingdom
- 22795/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials

