Community-scale models of microbiomes: Articulating metabolic modelling and metagenome sequencing
- PMID: 38243750
- PMCID: PMC10832553
- DOI: 10.1111/1751-7915.14396
Community-scale models of microbiomes: Articulating metabolic modelling and metagenome sequencing
Abstract
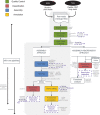
Building models is essential for understanding the functions and dynamics of microbial communities. Metabolic models built on genome-scale metabolic network reconstructions (GENREs) are especially relevant as a means to decipher the complex interactions occurring among species. Model reconstruction increasingly relies on metagenomics, which permits direct characterisation of naturally occurring communities that may contain organisms that cannot be isolated or cultured. In this review, we provide an overview of the field of metabolic modelling and its increasing reliance on and synergy with metagenomics and bioinformatics. We survey the means of assigning functions and reconstructing metabolic networks from (meta-)genomes, and present the variety and mathematical fundamentals of metabolic models that foster the understanding of microbial dynamics. We emphasise the characterisation of interactions and the scaling of model construction to large communities, two important bottlenecks in the applicability of these models. We give an overview of the current state of the art in metagenome sequencing and bioinformatics analysis, focusing on the reconstruction of genomes in microbial communities. Metagenomics benefits tremendously from third-generation sequencing, and we discuss the opportunities of long-read sequencing, strain-level characterisation and eukaryotic metagenomics. We aim at providing algorithmic and mathematical support, together with tool and application resources, that permit bridging the gap between metagenomics and metabolic modelling.
© 2024 The Authors. Microbial Biotechnology published by Applied Microbiology International and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

