The peroxisome: an update on mysteries 3.0
- PMID: 38244103
- PMCID: PMC10822820
- DOI: 10.1007/s00418-023-02259-5
The peroxisome: an update on mysteries 3.0
Abstract
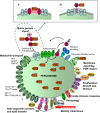
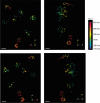
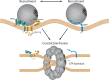
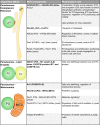
Peroxisomes are highly dynamic, oxidative organelles with key metabolic functions in cellular lipid metabolism, such as the β-oxidation of fatty acids and the synthesis of myelin sheath lipids, as well as the regulation of cellular redox balance. Loss of peroxisomal functions causes severe metabolic disorders in humans. Furthermore, peroxisomes also fulfil protective roles in pathogen and viral defence and immunity, highlighting their wider significance in human health and disease. This has sparked increasing interest in peroxisome biology and their physiological functions. This review presents an update and a continuation of three previous review articles addressing the unsolved mysteries of this remarkable organelle. We continue to highlight recent discoveries, advancements, and trends in peroxisome research, and address novel findings on the metabolic functions of peroxisomes, their biogenesis, protein import, membrane dynamics and division, as well as on peroxisome-organelle membrane contact sites and organelle cooperation. Furthermore, recent insights into peroxisome organisation through super-resolution microscopy are discussed. Finally, we address new roles for peroxisomes in immune and defence mechanisms and in human disorders, and for peroxisomal functions in different cell/tissue types, in particular their contribution to organ-specific pathologies.
Keywords: Membrane contact sites; Motility; Organelle biogenesis; Organelle division; Organelle dynamics; Peroxin; Peroxisome; Protein import; STED microscopy.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
- BB/W015420/1, BB/V018167/1, BB/T002255/1, BB/R016844/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/W015420/1, BB/V018167/1, BB/T002255/1, BB/R016844/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/W015420/1, BB/V018167/1, BB/T002255/1, BB/R016844/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources

