Functional genomics and small molecules in mitochondrial neurodevelopmental disorders
- PMID: 38244259
- PMCID: PMC10903096
- DOI: 10.1016/j.neurot.2024.e00316
Functional genomics and small molecules in mitochondrial neurodevelopmental disorders
Abstract
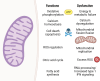
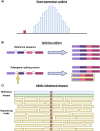
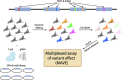
Mitochondria are critical for brain development and homeostasis. Therefore, pathogenic variation in the mitochondrial or nuclear genome which disrupts mitochondrial function frequently results in developmental disorders and neurodegeneration at the organismal level. Large-scale application of genome-wide technologies to individuals with mitochondrial diseases has dramatically accelerated identification of mitochondrial disease-gene associations in humans. Multi-omic and high-throughput studies involving transcriptomics, proteomics, metabolomics, and saturation genome editing are providing deeper insights into the functional consequence of mitochondrial genomic variation. Integration of deep phenotypic and genomic data through allelic series continues to uncover novel mitochondrial functions and permit mitochondrial gene function dissection on an unprecedented scale. Finally, mitochondrial disease-gene associations illuminate disease mechanisms and thereby direct therapeutic strategies involving small molecules and RNA-DNA therapeutics. This review summarizes progress in functional genomics and small molecule therapeutics in mitochondrial neurodevelopmental disorders.
Keywords: Functional genomics; Mitochondrial disease; Neurodevelopmental disorders; Small molecules; Therapeutics.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
The genetics of mitochondrial disease: dissecting mitochondrial pathology using multi-omic pipelines.J Pathol. 2021 Jul;254(4):430-442. doi: 10.1002/path.5641. Epub 2021 Mar 26. J Pathol. 2021. PMID: 33586140 Free PMC article. Review.
-
The Emerging Role of MitomiRs in the Pathophysiology of Human Disease.Adv Exp Med Biol. 2015;888:123-54. doi: 10.1007/978-3-319-22671-2_8. Adv Exp Med Biol. 2015. PMID: 26663182
-
Therapeutic potential of engineering the mitochondrial genome.Biochim Biophys Acta Mol Basis Dis. 2023 Oct;1869(7):166804. doi: 10.1016/j.bbadis.2023.166804. Epub 2023 Jul 8. Biochim Biophys Acta Mol Basis Dis. 2023. PMID: 37429560 Review.
-
The current landscape for the treatment of mitochondrial disorders.J Genet Genomics. 2018 Feb 20;45(2):71-77. doi: 10.1016/j.jgg.2017.11.008. Epub 2018 Feb 14. J Genet Genomics. 2018. PMID: 29502956 Review.
-
Mitochondrial Diseases: Hope for the Future.Cell. 2020 Apr 2;181(1):168-188. doi: 10.1016/j.cell.2020.02.051. Epub 2020 Mar 26. Cell. 2020. PMID: 32220313 Review.
Cited by
-
MST1, a novel therapeutic target for Alzheimer's disease, regulates mitochondrial homeostasis by mediating mitochondrial DNA transcription and the PI3K-Akt-ROS pathway.J Transl Med. 2024 Nov 22;22(1):1056. doi: 10.1186/s12967-024-05852-x. J Transl Med. 2024. PMID: 39578795 Free PMC article.
-
Mitochondrial disorders: Emerging paradigms and the road ahead to personalized medicine.Neurotherapeutics. 2024 Jan;21(1):e00332. doi: 10.1016/j.neurot.2024.e00332. Neurotherapeutics. 2024. PMID: 38355260 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

