UCHL1 is a potential molecular indicator and therapeutic target for neuroendocrine carcinomas
- PMID: 38244540
- PMCID: PMC10897521
- DOI: 10.1016/j.xcrm.2023.101381
UCHL1 is a potential molecular indicator and therapeutic target for neuroendocrine carcinomas
Abstract
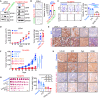
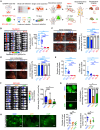
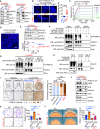
Neuroendocrine carcinomas, such as neuroendocrine prostate cancer and small-cell lung cancer, commonly have a poor prognosis and limited therapeutic options. We report that ubiquitin carboxy-terminal hydrolase L1 (UCHL1), a deubiquitinating enzyme, is elevated in tissues and plasma from patients with neuroendocrine carcinomas. Loss of UCHL1 decreases tumor growth and inhibits metastasis of these malignancies. UCHL1 maintains neuroendocrine differentiation and promotes cancer progression by regulating nucleoporin, POM121, and p53. UCHL1 binds, deubiquitinates, and stabilizes POM121 to regulate POM121-associated nuclear transport of E2F1 and c-MYC. Treatment with the UCHL1 inhibitor LDN-57444 slows tumor growth and metastasis across neuroendocrine carcinomas. The combination of UCHL1 inhibitors with cisplatin, the standard of care used for neuroendocrine carcinomas, significantly delays tumor growth in pre-clinical settings. Our study reveals mechanisms of UCHL1 function in regulating the progression of neuroendocrine carcinomas and identifies UCHL1 as a therapeutic target and potential molecular indicator for diagnosing and monitoring treatment responses in these malignancies.
Keywords: UCHL1; neuroblastoma; neuroendocrine carcinomas; neuroendocrine prostate cancer; nuclear pore complex; small cell lung cancer.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.C. is a consultant for Dotquant and received research funding under institutional SRAs from AbbVie, Astra Zeneca, Janssen Research, Gilead, Zenith Epigenetics, Forma Therapeutics, Bayer, Kronos, Foghorn, and MacroGenics. M.D. is a consultant for Regeneron, Beigene, Astra Zeneca, Sanofi/Genzyme, Eurofins, Janssen, and Genentech (uncompensated) and performs research at Merck, Genentech, CellSight, Novartis, AbbVie, United Therapeutics, Varian, Verily, and Celgene. J.H. is a consultant for or owns shares in Kingmed, MoreHealth, OptraScan, Genetron, Omnitura, Vetonco, York Biotechnology, Genecode, VIVA Biotech, and Sisu Pharma and received grants from Zenith Epigenetics, BioXcel Therapeutics, Inc., and Fortis Therapeutics. T.S. is a consultant for Dren Bio.
Figures



References
-
- Rindi G., Mete O., Uccella S., Basturk O., La Rosa S., Brosens L.A.A., Ezzat S., de Herder W.W., Klimstra D.S., Papotti M., Asa S.L. Overview of the 2022 WHO Classification of Neuroendocrine Neoplasms. Endocr. Pathol. 2022;33:115–154. - PubMed
-
- van Meerbeeck J.P., Fennell D.A., De Ruysscher D.K.M. Small-cell lung cancer. Lancet. 2011;378:1741–1755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

