BRCA2 promotes genomic integrity and therapy resistance primarily through its role in homology-directed repair
- PMID: 38244544
- PMCID: PMC11188060
- DOI: 10.1016/j.molcel.2023.12.025
BRCA2 promotes genomic integrity and therapy resistance primarily through its role in homology-directed repair
Abstract
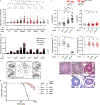
Tumor suppressor BRCA2 functions in homology-directed repair (HDR), the protection of stalled replication forks, and the suppression of replicative gaps, but their relative contributions to genome integrity and chemotherapy response are under scrutiny. Here, we report that mouse and human cells require a RAD51 filament stabilization motif in BRCA2 for fork protection and gap suppression but not HDR. In mice, the loss of fork protection/gap suppression does not compromise genome stability or shorten tumor latency. By contrast, HDR deficiency increases spontaneous and replication stress-induced chromosome aberrations and tumor predisposition. Unlike with HDR, fork protection/gap suppression defects are also observed in Brca2 heterozygous cells, likely due to reduced RAD51 stabilization at stalled forks/gaps. Gaps arise from PRIMPOL activity, which is associated with 5-hydroxymethyl-2'-deoxyuridine sensitivity due to the formation of SMUG1-generated abasic sites and is exacerbated by poly(ADP-ribose) polymerase (PARP) inhibition. However, HDR proficiency has the major role in mitigating sensitivity to chemotherapeutics, including PARP inhibitors.
Keywords: BRCA2; PARP inhibitors; PRIMPOL; RAD51; gap suppression; hmdU; homologous recombination; homology-directed repair; stalled fork protection.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
BRCA2 promotes genomic integrity and therapy resistance primarily through its role in homology-directed repair.bioRxiv [Preprint]. 2023 Apr 11:2023.04.11.536470. doi: 10.1101/2023.04.11.536470. bioRxiv. 2023. Update in: Mol Cell. 2024 Feb 1;84(3):447-462.e10. doi: 10.1016/j.molcel.2023.12.025. PMID: 37090587 Free PMC article. Updated. Preprint.
References
-
- Pennington KP, Walsh T, Harrell MI, Lee MK, Pennil CC, Rendi MH, Thornton A, Norquist BM, Casadei S, Nord AS, et al. (2014). Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res 20, 764–775. 10.1158/1078-0432.CCR-13-2287. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

