Cell state dependent effects of Bmal1 on melanoma immunity and tumorigenicity
- PMID: 38245503
- PMCID: PMC10799901
- DOI: 10.1038/s41467-024-44778-2
Cell state dependent effects of Bmal1 on melanoma immunity and tumorigenicity
Abstract
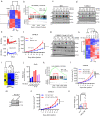
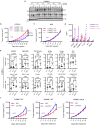
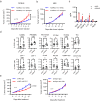
The circadian clock regulator Bmal1 modulates tumorigenesis, but its reported effects are inconsistent. Here, we show that Bmal1 has a context-dependent role in mouse melanoma tumor growth. Loss of Bmal1 in YUMM2.1 or B16-F10 melanoma cells eliminates clock function and diminishes hypoxic gene expression and tumorigenesis, which could be rescued by ectopic expression of HIF1α in YUMM2.1 cells. By contrast, over-expressed wild-type or a transcriptionally inactive mutant Bmal1 non-canonically sequester myosin heavy chain 9 (Myh9) to increase MRTF-SRF activity and AP-1 transcriptional signature, and shift YUMM2.1 cells from a Sox10high to a Sox9high immune resistant, mesenchymal cell state that is found in human melanomas. Our work describes a link between Bmal1, Myh9, mouse melanoma cell plasticity, and tumor immunity. This connection may underlie cancer therapeutic resistance and underpin the link between the circadian clock, MRTF-SRF and the cytoskeleton.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Sancar, A. & Van Gelder, R.N. Clocks, cancer, and chronochemotherapy. Science371, eabb0738 (2021). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

