Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH
- PMID: 38245551
- PMCID: PMC10799931
- DOI: 10.1038/s41467-024-44879-y
Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH
Abstract
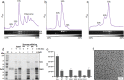
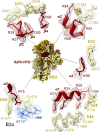
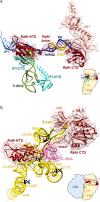
Ribosome hibernation is a key survival strategy bacteria adopt under environmental stress, where a protein, hibernation promotion factor (HPF), transitorily inactivates the ribosome. Mycobacterium tuberculosis encounters hypoxia (low oxygen) as a major stress in the host macrophages, and upregulates the expression of RafH protein, which is crucial for its survival. The RafH, a dual domain HPF, an orthologue of bacterial long HPF (HPFlong), hibernates ribosome in 70S monosome form, whereas in other bacteria, the HPFlong induces 70S ribosome dimerization and hibernates its ribosome in 100S disome form. Here, we report the cryo- EM structure of M. smegmatis, a close homolog of M. tuberculosis, 70S ribosome in complex with the RafH factor at an overall 2.8 Å resolution. The N- terminus domain (NTD) of RafH binds to the decoding center, similarly to HPFlong NTD. In contrast, the C- terminus domain (CTD) of RafH, which is larger than the HPFlong CTD, binds to a distinct site at the platform binding center of the ribosomal small subunit. The two domain-connecting linker regions, which remain mostly disordered in earlier reported HPFlong structures, interact mainly with the anti-Shine Dalgarno sequence of the 16S rRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

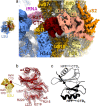

Similar articles
-
A general mechanism of ribosome dimerization revealed by single-particle cryo-electron microscopy.Nat Commun. 2017 Sep 28;8(1):722. doi: 10.1038/s41467-017-00718-x. Nat Commun. 2017. PMID: 28959045 Free PMC article.
-
Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1.Nat Microbiol. 2018 Oct;3(10):1115-1121. doi: 10.1038/s41564-018-0237-0. Epub 2018 Sep 3. Nat Microbiol. 2018. PMID: 30177741
-
How hibernation factors RMF, HPF, and YfiA turn off protein synthesis.Science. 2012 May 18;336(6083):915-8. doi: 10.1126/science.1218538. Science. 2012. PMID: 22605777 Free PMC article.
-
Unique structural features of the Mycobacterium ribosome.Prog Biophys Mol Biol. 2020 May;152:15-24. doi: 10.1016/j.pbiomolbio.2019.12.001. Epub 2019 Dec 10. Prog Biophys Mol Biol. 2020. PMID: 31858996 Review.
-
Survival of the drowsiest: the hibernating 100S ribosome in bacterial stress management.Curr Genet. 2018 Aug;64(4):753-760. doi: 10.1007/s00294-017-0796-2. Epub 2017 Dec 14. Curr Genet. 2018. PMID: 29243175 Free PMC article. Review.
Cited by
-
Methotrimeprazine is a neuroprotective antiviral in JEV infection via adaptive ER stress and autophagy.EMBO Mol Med. 2024 Jan;16(1):185-217. doi: 10.1038/s44321-023-00014-w. Epub 2024 Jan 2. EMBO Mol Med. 2024. PMID: 38177535 Free PMC article.
-
Staphylococcal exoribonuclease YhaM destabilizes ribosomes by targeting the mRNA of a hibernation factor.Nucleic Acids Res. 2024 Aug 27;52(15):8998-9013. doi: 10.1093/nar/gkae596. Nucleic Acids Res. 2024. PMID: 38979572 Free PMC article.
-
Novel archaeal ribosome dimerization factor facilitating unique 30S-30S dimerization.Nucleic Acids Res. 2025 Jan 11;53(2):gkae1324. doi: 10.1093/nar/gkae1324. Nucleic Acids Res. 2025. PMID: 39797736 Free PMC article.
-
Rippling life on a dormant planet: hibernation of ribosomes, RNA polymerases, and other essential enzymes.Front Microbiol. 2024 May 6;15:1386179. doi: 10.3389/fmicb.2024.1386179. eCollection 2024. Front Microbiol. 2024. PMID: 38770025 Free PMC article. Review.
References
-
- Bremer, H. & Dennis, P. P. Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus 3, 10–1128 (2008). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

